 View on TensorFlow.org View on TensorFlow.org
|
 Run in Google Colab Run in Google Colab
|
 View source on GitHub View source on GitHub
|
 Download notebook Download notebook
|
Overview
This tutorial shows how to use tfio.image.decode_dicom_image in TensorFlow IO to decode DICOM files with TensorFlow.
Setup and Usage
Download DICOM image
The DICOM image used in this tutorial is from the NIH Chest X-ray dataset.
The NIH Chest X-ray dataset consists of 100,000 de-identified images of chest x-rays in PNG format, provided by NIH Clinical Center and could be downloaded through this link.
Google Cloud also provides a DICOM version of the images, available in Cloud Storage.
In this tutorial, you will download a sample file of the dataset from the GitHub repo
- Xiaosong Wang, Yifan Peng, Le Lu, Zhiyong Lu, Mohammadhadi Bagheri, Ronald Summers, ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases, IEEE CVPR, pp. 3462-3471, 2017
curl -OL https://github.com/tensorflow/io/raw/master/docs/tutorials/dicom/dicom_00000001_000.dcmls -l dicom_00000001_000.dcm
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
100 164 0 164 0 0 600 0 --:--:-- --:--:-- --:--:-- 598
100 1024k 100 1024k 0 0 1915k 0 --:--:-- --:--:-- --:--:-- 1915k
-rw-rw-r-- 1 kbuilder kokoro 1049332 Nov 22 03:47 dicom_00000001_000.dcm
Install required Packages, and restart runtime
try:
# Use the Colab's preinstalled TensorFlow 2.x
%tensorflow_version 2.x
except:
pass
pip install tensorflow-ioDecode DICOM image
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
import tensorflow_io as tfio
image_bytes = tf.io.read_file('dicom_00000001_000.dcm')
image = tfio.image.decode_dicom_image(image_bytes, dtype=tf.uint16)
skipped = tfio.image.decode_dicom_image(image_bytes, on_error='skip', dtype=tf.uint8)
lossy_image = tfio.image.decode_dicom_image(image_bytes, scale='auto', on_error='lossy', dtype=tf.uint8)
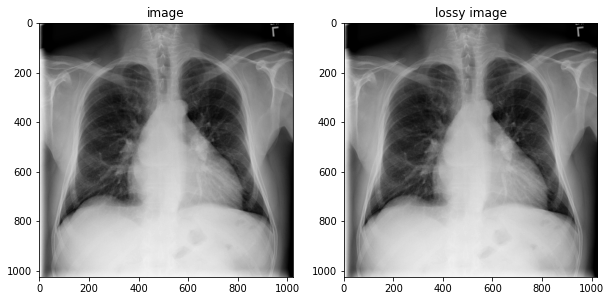
fig, axes = plt.subplots(1,2, figsize=(10,10))
axes[0].imshow(np.squeeze(image.numpy()), cmap='gray')
axes[0].set_title('image')
axes[1].imshow(np.squeeze(lossy_image.numpy()), cmap='gray')
axes[1].set_title('lossy image');
2021-11-22 03:47:53.016507: E tensorflow/stream_executor/cuda/cuda_driver.cc:271] failed call to cuInit: CUDA_ERROR_NO_DEVICE: no CUDA-capable device is detected
Decode DICOM Metadata and working with Tags
decode_dicom_data decodes tag information. dicom_tags contains useful information as the patient's age and sex, so you can use DICOM tags such as dicom_tags.PatientsAge and dicom_tags.PatientsSex. tensorflow_io borrow the same tag notation from the pydicom dicom package.
tag_id = tfio.image.dicom_tags.PatientsAge
tag_value = tfio.image.decode_dicom_data(image_bytes,tag_id)
print(tag_value)
tf.Tensor(b'58', shape=(), dtype=string)
print(f"PatientsAge : {tag_value.numpy().decode('UTF-8')}")
PatientsAge : 58
tag_id = tfio.image.dicom_tags.PatientsSex
tag_value = tfio.image.decode_dicom_data(image_bytes,tag_id)
print(f"PatientsSex : {tag_value.numpy().decode('UTF-8')}")
PatientsSex : M
