 View on TensorFlow.org View on TensorFlow.org
|
 Run in Google Colab Run in Google Colab
|
 View on GitHub View on GitHub
|
 Download notebook Download notebook
|
Welcome to the Learning to Rank Colab for TensorFlow Decision Forests (TF-DF). In this colab, you will learn how to use TF-DF for ranking.
This colab assumes you are familiar with the concepts presented the Beginner colab, notably about the installation about TF-DF.
In this colab, you will:
- Learn what a ranking model is.
- Train a Gradient Boosted Trees models on the LETOR3 dataset.
- Evaluate the quality of this model.
Installing TensorFlow Decision Forests
Install TF-DF by running the following cell.
pip install tensorflow_decision_forestsWurlitzer is needed to display the detailed training logs in Colabs (when using verbose=2 in the model constructor).
pip install wurlitzerImporting libraries
import os
# Keep using Keras 2
os.environ['TF_USE_LEGACY_KERAS'] = '1'
import tensorflow_decision_forests as tfdf
import numpy as np
import pandas as pd
import tensorflow as tf
import tf_keras
import math
2026-01-12 14:14:58.435612: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:467] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered WARNING: All log messages before absl::InitializeLog() is called are written to STDERR E0000 00:00:1768227298.458012 158192 cuda_dnn.cc:8579] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered E0000 00:00:1768227298.465464 158192 cuda_blas.cc:1407] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered W0000 00:00:1768227298.483480 158192 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once. W0000 00:00:1768227298.483499 158192 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once. W0000 00:00:1768227298.483502 158192 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once. W0000 00:00:1768227298.483504 158192 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
The hidden code cell limits the output height in colab.
from IPython.core.magic import register_line_magic
from IPython.display import Javascript
from IPython.display import display as ipy_display
# Some of the model training logs can cover the full
# screen if not compressed to a smaller viewport.
# This magic allows setting a max height for a cell.
@register_line_magic
def set_cell_height(size):
ipy_display(
Javascript("google.colab.output.setIframeHeight(0, true, {maxHeight: " +
str(size) + "})"))
# Check the version of TensorFlow Decision Forests
print("Found TensorFlow Decision Forests v" + tfdf.__version__)
Found TensorFlow Decision Forests v1.12.0
What is a ranking model?
The goal of a ranking model is to correctly order items. For example, ranking can be used to select the best documents to retrieve following a user query.
A common way to represent a Ranking dataset is with a "relevance" score: The order of the elements is defined by their relevance: Items of greater relevance should be before lower relevance items. The cost of a mistake is defined by the difference between the relevance of the predicted item with the relevance of the correct item. For example, misordering two items with respective relevance 3 and 4 is not as bad as misordering two items with respective relevance 1 and 5.
TF-DF expects ranking datasets to be presented in a "flat" format. A dataset of queries and corresponding documents might look like this:
| query | document_id | feature_1 | feature_2 | relevance |
|---|---|---|---|---|
| cat | 1 | 0.1 | blue | 4 |
| cat | 2 | 0.5 | green | 1 |
| cat | 3 | 0.2 | red | 2 |
| dog | 4 | NA | red | 0 |
| dog | 5 | 0.2 | red | 0 |
| dog | 6 | 0.6 | green | 1 |
The relevance/label is a floating point numerical value between 0 and 5 (generally between 0 and 4) where 0 means "completely unrelated", 4 means "very relevant" and 5 means "same as the query".
In this example, Document 1 is very relevant to the query "cat", while document 2 is only "related" to cats. There are no documents is really talking about "dog" (the highest relevance is 1 for the document 6). However, the dog query is still expecting to return document 6 (since this is the document that talks the "most" about dogs).
Interestingly, decision forests are often good rankers, and many state-of-the-art ranking models are decision forests.
Let's train a Ranking model
In this example, use a sample of the
LETOR3
dataset. More precisely, we want to download the OHSUMED.zip from the LETOR3 repo. This dataset is stored in the
libsvm format, so we will need to convert it to csv.
archive_path = tf_keras.utils.get_file("letor.zip",
"https://download.microsoft.com/download/E/7/E/E7EABEF1-4C7B-4E31-ACE5-73927950ED5E/Letor.zip",
extract=True)
# Path to a ranking ataset using libsvm format.
raw_dataset_path = os.path.join(os.path.dirname(archive_path),"OHSUMED/Data/Fold1/trainingset.txt")
Downloading data from https://download.microsoft.com/download/E/7/E/E7EABEF1-4C7B-4E31-ACE5-73927950ED5E/Letor.zip 61824018/61824018 [==============================] - 4s 0us/step
Here are the first lines of the dataset:
head {raw_dataset_path}The first step is to convert this dataset to the "flat" format mentioned above.
def convert_libsvm_to_csv(src_path, dst_path):
"""Converts a libsvm ranking dataset into a flat csv file.
Note: This code is specific to the LETOR3 dataset.
"""
dst_handle = open(dst_path, "w")
first_line = True
for src_line in open(src_path,"r"):
# Note: The last 3 items are comments.
items = src_line.split(" ")[:-3]
relevance = items[0]
group = items[1].split(":")[1]
features = [ item.split(":") for item in items[2:]]
if first_line:
# Csv header
dst_handle.write("relevance,group," + ",".join(["f_" + feature[0] for feature in features]) + "\n")
first_line = False
dst_handle.write(relevance + ",g_" + group + "," + (",".join([feature[1] for feature in features])) + "\n")
dst_handle.close()
# Convert the dataset.
csv_dataset_path="/tmp/ohsumed.csv"
convert_libsvm_to_csv(raw_dataset_path, csv_dataset_path)
# Load a dataset into a Pandas Dataframe.
dataset_df = pd.read_csv(csv_dataset_path)
# Display the first 3 examples.
dataset_df.head(3)
In this dataset, each row represents a pair of query/document (called "group"). The "relevance" tells how much the query matches the document.
The features of the query and the document are merged together in "f1-25". The exact definition of the features is not known, but it would be something like:
- Number of words in queries
- Number of common words between the query and the document
- Cosinus similarity between an embedding of the query and an embedding of the document.
- ...
Let's convert the Pandas Dataframe into a TensorFlow Dataset:
dataset_ds = tfdf.keras.pd_dataframe_to_tf_dataset(dataset_df, label="relevance", task=tfdf.keras.Task.RANKING)
I0000 00:00:1768227309.575367 158192 gpu_device.cc:2019] Created device /job:localhost/replica:0/task:0/device:GPU:0 with 13638 MB memory: -> device: 0, name: Tesla T4, pci bus id: 0000:00:05.0, compute capability: 7.5 I0000 00:00:1768227309.577633 158192 gpu_device.cc:2019] Created device /job:localhost/replica:0/task:0/device:GPU:1 with 13756 MB memory: -> device: 1, name: Tesla T4, pci bus id: 0000:00:06.0, compute capability: 7.5 I0000 00:00:1768227309.579812 158192 gpu_device.cc:2019] Created device /job:localhost/replica:0/task:0/device:GPU:2 with 13756 MB memory: -> device: 2, name: Tesla T4, pci bus id: 0000:00:07.0, compute capability: 7.5 I0000 00:00:1768227309.581958 158192 gpu_device.cc:2019] Created device /job:localhost/replica:0/task:0/device:GPU:3 with 13756 MB memory: -> device: 3, name: Tesla T4, pci bus id: 0000:00:08.0, compute capability: 7.5
Let's configure and train our Ranking model.
%set_cell_height 400
model = tfdf.keras.GradientBoostedTreesModel(
task=tfdf.keras.Task.RANKING,
ranking_group="group",
num_trees=50)
model.fit(dataset_ds)
<IPython.core.display.Javascript object>
Warning: The `num_threads` constructor argument is not set and the number of CPU is os.cpu_count()=32 > 32. Setting num_threads to 32. Set num_threads manually to use more than 32 cpus.
WARNING:absl:The `num_threads` constructor argument is not set and the number of CPU is os.cpu_count()=32 > 32. Setting num_threads to 32. Set num_threads manually to use more than 32 cpus.
Use /tmpfs/tmp/tmpc9wak2d3 as temporary training directory
Reading training dataset...
WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
W0000 00:00:1768227309.968180 158192 gradient_boosted_trees.cc:1873] "goss_alpha" set but "sampling_method" not equal to "GOSS".
W0000 00:00:1768227309.968237 158192 gradient_boosted_trees.cc:1883] "goss_beta" set but "sampling_method" not equal to "GOSS".
W0000 00:00:1768227309.968240 158192 gradient_boosted_trees.cc:1897] "selective_gradient_boosting_ratio" set but "sampling_method" not equal to "SELGB".
Training dataset read in 0:00:03.864118. Found 9219 examples.
Training model...
I0000 00:00:1768227313.858225 158192 kernel.cc:782] Start Yggdrasil model training
I0000 00:00:1768227313.858260 158192 kernel.cc:783] Collect training examples
I0000 00:00:1768227313.858268 158192 kernel.cc:795] Dataspec guide:
column_guides {
column_name_pattern: "^__LABEL$"
type: NUMERICAL
}
default_column_guide {
categorial {
max_vocab_count: 2000
}
discretized_numerical {
maximum_num_bins: 255
}
}
ignore_columns_without_guides: false
detect_numerical_as_discretized_numerical: false
I0000 00:00:1768227313.858676 158192 kernel.cc:401] Number of batches: 10
I0000 00:00:1768227313.858694 158192 kernel.cc:402] Number of examples: 9219
I0000 00:00:1768227313.860506 158192 kernel.cc:802] Training dataset:
Number of records: 9219
Number of columns: 27
Number of columns by type:
NUMERICAL: 26 (96.2963%)
HASH: 1 (3.7037%)
Columns:
NUMERICAL: 26 (96.2963%)
0: "__LABEL" NUMERICAL mean:0.495607 min:0 max:2 sd:0.744403
1: "f_1" NUMERICAL mean:1.21141 min:0 max:7 sd:1.15164
2: "f_10" NUMERICAL mean:4.20167 min:0 max:21.0369 sd:3.88154
3: "f_11" NUMERICAL mean:4.33312 min:0 max:59 sd:4.67348
4: "f_12" NUMERICAL mean:1.91775 min:0 max:9.75731 sd:1.61639
5: "f_13" NUMERICAL mean:0.0457776 min:0 max:0.384615 sd:0.0466109
6: "f_14" NUMERICAL mean:0.0447853 min:0 max:0.361682 sd:0.0451178
7: "f_15" NUMERICAL mean:21.3512 min:11.4845 max:39.1502 sd:6.03344
8: "f_16" NUMERICAL mean:6.70697 min:3.95484 max:12.369 sd:1.80357
9: "f_17" NUMERICAL mean:19.534 min:10.2355 max:40.1808 sd:6.08569
10: "f_18" NUMERICAL mean:0.195288 min:0 max:1.3212 sd:0.187686
11: "f_19" NUMERICAL mean:20.1237 min:0 max:176.805 sd:20.9556
12: "f_2" NUMERICAL mean:0.825689 min:0 max:4.27667 sd:0.772347
13: "f_20" NUMERICAL mean:1.8782 min:0 max:11.6585 sd:1.75127
14: "f_21" NUMERICAL mean:12.2408 min:3.18098 max:45.0899 sd:6.76927
15: "f_22" NUMERICAL mean:2.31505 min:1.15719 max:3.80866 sd:0.666958
16: "f_23" NUMERICAL mean:-6.0857 min:-9.49097 max:-1.85651 sd:2.13886
17: "f_24" NUMERICAL mean:-5.83816 min:-9.22971 max:-1.02685 sd:1.96046
18: "f_25" NUMERICAL mean:-5.98972 min:-9.60073 max:-1.02685 sd:2.16203
19: "f_3" NUMERICAL mean:0.161865 min:0 max:1 sd:0.165642
20: "f_4" NUMERICAL mean:0.149731 min:0 max:0.892574 sd:0.149804
21: "f_5" NUMERICAL mean:26.3233 min:15.3432 max:51.3862 sd:7.40231
22: "f_6" NUMERICAL mean:7.82971 min:4.24645 max:15.3248 sd:2.09136
23: "f_7" NUMERICAL mean:26.9268 min:15.3265 max:52.0258 sd:8.07986
24: "f_8" NUMERICAL mean:0.64629 min:0 max:3.59024 sd:0.614985
25: "f_9" NUMERICAL mean:6.78251 min:0 max:47.7046 sd:6.26551
HASH: 1 (3.7037%)
26: "group" HASH
Terminology:
nas: Number of non-available (i.e. missing) values.
ood: Out of dictionary.
manually-defined: Attribute whose type is manually defined by the user, i.e., the type was not automatically inferred.
tokenized: The attribute value is obtained through tokenization.
has-dict: The attribute is attached to a string dictionary e.g. a categorical attribute stored as a string.
vocab-size: Number of unique values.
I0000 00:00:1768227313.860548 158192 kernel.cc:818] Configure learner
W0000 00:00:1768227313.860782 158192 gradient_boosted_trees.cc:1873] "goss_alpha" set but "sampling_method" not equal to "GOSS".
W0000 00:00:1768227313.860795 158192 gradient_boosted_trees.cc:1883] "goss_beta" set but "sampling_method" not equal to "GOSS".
W0000 00:00:1768227313.860798 158192 gradient_boosted_trees.cc:1897] "selective_gradient_boosting_ratio" set but "sampling_method" not equal to "SELGB".
I0000 00:00:1768227313.860844 158192 kernel.cc:831] Training config:
learner: "GRADIENT_BOOSTED_TREES"
features: "^f_1$"
features: "^f_10$"
features: "^f_11$"
features: "^f_12$"
features: "^f_13$"
features: "^f_14$"
features: "^f_15$"
features: "^f_16$"
features: "^f_17$"
features: "^f_18$"
features: "^f_19$"
features: "^f_2$"
features: "^f_20$"
features: "^f_21$"
features: "^f_22$"
features: "^f_23$"
features: "^f_24$"
features: "^f_25$"
features: "^f_3$"
features: "^f_4$"
features: "^f_5$"
features: "^f_6$"
features: "^f_7$"
features: "^f_8$"
features: "^f_9$"
label: "^__LABEL$"
task: RANKING
random_seed: 123456
ranking_group: "group"
metadata {
framework: "TF Keras"
}
pure_serving_model: false
[yggdrasil_decision_forests.model.gradient_boosted_trees.proto.gradient_boosted_trees_config] {
num_trees: 50
decision_tree {
max_depth: 6
min_examples: 5
in_split_min_examples_check: true
keep_non_leaf_label_distribution: true
num_candidate_attributes: -1
missing_value_policy: GLOBAL_IMPUTATION
allow_na_conditions: false
categorical_set_greedy_forward {
sampling: 0.1
max_num_items: -1
min_item_frequency: 1
}
growing_strategy_local {
}
categorical {
cart {
}
}
axis_aligned_split {
}
internal {
sorting_strategy: PRESORTED
}
uplift {
min_examples_in_treatment: 5
split_score: KULLBACK_LEIBLER
}
numerical_vector_sequence {
max_num_test_examples: 1000
num_random_selected_anchors: 100
}
}
shrinkage: 0.1
loss: DEFAULT
validation_set_ratio: 0.1
validation_interval_in_trees: 1
early_stopping: VALIDATION_LOSS_INCREASE
early_stopping_num_trees_look_ahead: 30
l2_regularization: 0
lambda_loss: 1
mart {
}
adapt_subsample_for_maximum_training_duration: false
l1_regularization: 0
use_hessian_gain: false
l2_regularization_categorical: 1
xe_ndcg {
ndcg_truncation: 5
}
stochastic_gradient_boosting {
ratio: 1
}
apply_link_function: true
compute_permutation_variable_importance: false
early_stopping_initial_iteration: 10
}
I0000 00:00:1768227313.861218 158192 kernel.cc:834] Deployment config:
cache_path: "/tmpfs/tmp/tmpc9wak2d3/working_cache"
num_threads: 32
try_resume_training: true
I0000 00:00:1768227313.861467 158420 kernel.cc:895] Train model
I0000 00:00:1768227313.861639 158420 gradient_boosted_trees.cc:577] Default loss set to LAMBDA_MART_NDCG
I0000 00:00:1768227313.861666 158420 gradient_boosted_trees.cc:1190] Training gradient boosted tree on 9219 example(s) and 25 feature(s).
I0000 00:00:1768227313.861893 158420 gradient_boosted_trees.cc:2707] Split training/validation dataset by "group". 63 groups found in 9219 examples i.e. 146.333 examples/groups.
I0000 00:00:1768227313.863428 158420 gradient_boosted_trees.cc:1230] 8365 examples used for training and 854 examples used for validation
I0000 00:00:1768227313.864090 158420 loss_interface.cc:139] Found 58 groups in 8365 examples.
I0000 00:00:1768227313.864167 158420 loss_interface.cc:139] Found 5 groups in 854 examples.
I0000 00:00:1768227313.870274 158420 gpu.cc:93] Cannot initialize GPU: Not compiled with GPU support
I0000 00:00:1768227313.884998 158420 gradient_boosted_trees.cc:1632] Train tree 1/50 train-loss:-0.346669 train-NDCG@5:0.346669 valid-loss:-0.262935 valid-NDCG@5:0.262935 [total:0.01s iter:0.01s]
I0000 00:00:1768227313.898332 158420 gradient_boosted_trees.cc:1632] Train tree 2/50 train-loss:-0.412635 train-NDCG@5:0.412635 valid-loss:-0.335301 valid-NDCG@5:0.335301 [total:0.03s iter:0.01s]
I0000 00:00:1768227313.912874 158420 gradient_boosted_trees.cc:1634] Train tree 3/50 train-loss:-0.468270 train-NDCG@5:0.468270 valid-loss:-0.341295 valid-NDCG@5:0.341295 [total:0.04s iter:0.01s]
Model trained in 0:00:00.624905
Compiling model...
I0000 00:00:1768227314.460889 158420 early_stopping.cc:54] Early stop of the training because the validation loss does not decrease anymore. Best valid-loss: -0.438692
I0000 00:00:1768227314.460950 158420 gradient_boosted_trees.cc:1669] Create final snapshot of the model at iteration 41
I0000 00:00:1768227314.462473 158420 gradient_boosted_trees.cc:279] Truncates the model to 12 tree(s) i.e. 12 iteration(s).
I0000 00:00:1768227314.462743 158420 gradient_boosted_trees.cc:341] Final model num-trees:12 valid-loss:-0.438692 valid-NDCG@5:0.438692
I0000 00:00:1768227314.463210 158420 kernel.cc:926] Export model in log directory: /tmpfs/tmp/tmpc9wak2d3 with prefix 6df97c3231804358
I0000 00:00:1768227314.464330 158420 kernel.cc:944] Save model in resources
I0000 00:00:1768227314.467654 158192 abstract_model.cc:921] Model self evaluation:
Task: RANKING
Loss (LAMBDA_MART_NDCG@5): -0.438692
NDCG@5: 0.438692
MRR@0: 0
Precision@1: 0
Default NDCG@5: 0
Number of groups: 0
Number of items in groups: mean:0 min:0 max:0
WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
I0000 00:00:1768227314.478268 158192 quick_scorer_extended.cc:927] The binary was compiled without AVX2 support, but your CPU supports it. Enable it for faster model inference.
I0000 00:00:1768227314.478349 158192 abstract_model.cc:1439] Engine "GradientBoostedTreesQuickScorerExtended" built
Model compiled.
<tf_keras.src.callbacks.History at 0x7f2ae657bac0>
We can now look at the quality of the model on the validation dataset. By default, TF-DF trains ranking models to optimize the NDCG. The NDCG is a value between 0 and 1, where 1 is the perfect score. For this reason, -NDCG is the model loss.
import matplotlib.pyplot as plt
logs = model.make_inspector().training_logs()
plt.figure(figsize=(12, 4))
plt.subplot(1, 2, 1)
plt.plot([log.num_trees for log in logs], [log.evaluation.ndcg for log in logs])
plt.xlabel("Number of trees")
plt.ylabel("NDCG (validation)")
plt.subplot(1, 2, 2)
plt.plot([log.num_trees for log in logs], [log.evaluation.loss for log in logs])
plt.xlabel("Number of trees")
plt.ylabel("Loss (validation)")
plt.show()
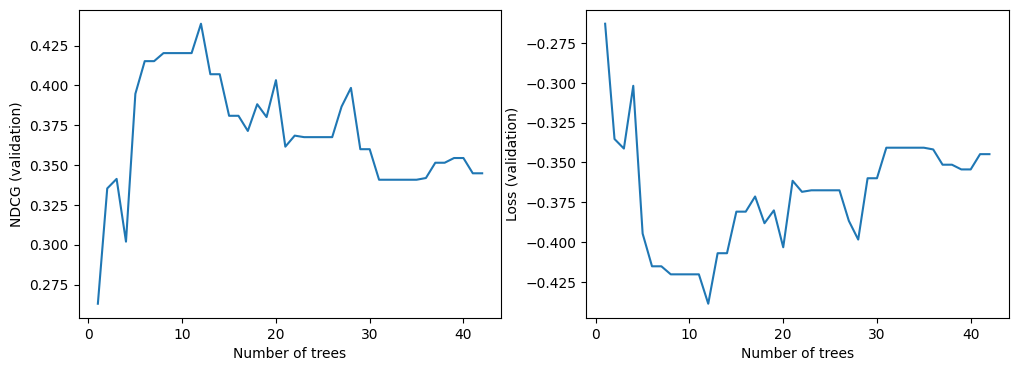
As for all TF-DF models, you can also look at the model report (Note: The model report also contains the training logs):
%set_cell_height 400
model.summary()
<IPython.core.display.Javascript object>
Model: "gradient_boosted_trees_model"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
=================================================================
Total params: 1 (1.00 Byte)
Trainable params: 0 (0.00 Byte)
Non-trainable params: 1 (1.00 Byte)
_________________________________________________________________
Type: "GRADIENT_BOOSTED_TREES"
Task: RANKING
Label: "__LABEL"
Rank group: "group"
Input Features (25):
f_1
f_10
f_11
f_12
f_13
f_14
f_15
f_16
f_17
f_18
f_19
f_2
f_20
f_21
f_22
f_23
f_24
f_25
f_3
f_4
f_5
f_6
f_7
f_8
f_9
No weights
Variable Importance: INV_MEAN_MIN_DEPTH:
1. "f_9" 0.326164 ################
2. "f_3" 0.318071 ###############
3. "f_8" 0.308922 #############
4. "f_4" 0.271175 #########
5. "f_19" 0.221570 ###
6. "f_10" 0.215666 ##
7. "f_11" 0.206509 #
8. "f_22" 0.204742 #
9. "f_25" 0.204497 #
10. "f_23" 0.203238
11. "f_21" 0.200830
12. "f_24" 0.200445
13. "f_12" 0.198840
14. "f_18" 0.197676
15. "f_20" 0.196634
16. "f_6" 0.196085
17. "f_16" 0.196061
18. "f_2" 0.195683
19. "f_5" 0.195683
20. "f_13" 0.195559
21. "f_17" 0.195559
Variable Importance: NUM_AS_ROOT:
1. "f_3" 4.000000 ################
2. "f_4" 4.000000 ################
3. "f_8" 3.000000 ##########
4. "f_9" 1.000000
Variable Importance: NUM_NODES:
1. "f_8" 25.000000 ################
2. "f_19" 18.000000 ###########
3. "f_10" 15.000000 #########
4. "f_9" 14.000000 ########
5. "f_3" 13.000000 ########
6. "f_23" 7.000000 ####
7. "f_24" 6.000000 ###
8. "f_11" 5.000000 ##
9. "f_21" 5.000000 ##
10. "f_25" 5.000000 ##
11. "f_4" 5.000000 ##
12. "f_22" 4.000000 ##
13. "f_12" 3.000000 #
14. "f_20" 3.000000 #
15. "f_16" 2.000000
16. "f_6" 2.000000
17. "f_13" 1.000000
18. "f_17" 1.000000
19. "f_18" 1.000000
20. "f_2" 1.000000
21. "f_5" 1.000000
Variable Importance: SUM_SCORE:
1. "f_8" 10779.340861 ################
2. "f_9" 8831.772410 #############
3. "f_3" 4526.101184 ######
4. "f_4" 4360.245403 ######
5. "f_19" 2325.288894 ###
6. "f_10" 1881.848369 ##
7. "f_21" 1674.980191 ##
8. "f_11" 1127.632256 #
9. "f_23" 1021.834252 #
10. "f_24" 914.851512 #
11. "f_22" 885.619576 #
12. "f_25" 748.665007 #
13. "f_20" 310.610858
14. "f_16" 298.972842
15. "f_6" 212.376573
16. "f_12" 130.725240
17. "f_2" 112.124991
18. "f_18" 86.341193
19. "f_5" 65.103908
20. "f_13" 57.966947
21. "f_17" 21.930388
Loss: LAMBDA_MART_NDCG@5
Validation loss value: -0.438692
Number of trees per iteration: 1
Node format: NOT_SET
Number of trees: 12
Total number of nodes: 286
Number of nodes by tree:
Count: 12 Average: 23.8333 StdDev: 3.50793
Min: 17 Max: 29 Ignored: 0
----------------------------------------------
[ 17, 18) 1 8.33% 8.33% ###
[ 18, 19) 0 0.00% 8.33%
[ 19, 20) 1 8.33% 16.67% ###
[ 20, 21) 0 0.00% 16.67%
[ 21, 22) 2 16.67% 33.33% #######
[ 22, 23) 0 0.00% 33.33%
[ 23, 24) 1 8.33% 41.67% ###
[ 24, 25) 0 0.00% 41.67%
[ 25, 26) 3 25.00% 66.67% ##########
[ 26, 27) 0 0.00% 66.67%
[ 27, 28) 3 25.00% 91.67% ##########
[ 28, 29) 0 0.00% 91.67%
[ 29, 29] 1 8.33% 100.00% ###
Depth by leafs:
Count: 149 Average: 4.14094 StdDev: 1.08696
Min: 1 Max: 5 Ignored: 0
----------------------------------------------
[ 1, 2) 2 1.34% 1.34%
[ 2, 3) 18 12.08% 13.42% ##
[ 3, 4) 13 8.72% 22.15% ##
[ 4, 5) 40 26.85% 48.99% #####
[ 5, 5] 76 51.01% 100.00% ##########
Number of training obs by leaf:
Count: 149 Average: 673.691 StdDev: 2015.44
Min: 5 Max: 8211 Ignored: 0
----------------------------------------------
[ 5, 415) 127 85.23% 85.23% ##########
[ 415, 825) 6 4.03% 89.26%
[ 825, 1236) 2 1.34% 90.60%
[ 1236, 1646) 0 0.00% 90.60%
[ 1646, 2056) 0 0.00% 90.60%
[ 2056, 2467) 1 0.67% 91.28%
[ 2467, 2877) 0 0.00% 91.28%
[ 2877, 3287) 0 0.00% 91.28%
[ 3287, 3698) 1 0.67% 91.95%
[ 3698, 4108) 0 0.00% 91.95%
[ 4108, 4518) 0 0.00% 91.95%
[ 4518, 4929) 1 0.67% 92.62%
[ 4929, 5339) 0 0.00% 92.62%
[ 5339, 5749) 0 0.00% 92.62%
[ 5749, 6160) 1 0.67% 93.29%
[ 6160, 6570) 0 0.00% 93.29%
[ 6570, 6980) 0 0.00% 93.29%
[ 6980, 7391) 0 0.00% 93.29%
[ 7391, 7801) 8 5.37% 98.66% #
[ 7801, 8211] 2 1.34% 100.00%
Attribute in nodes:
25 : f_8 [NUMERICAL]
18 : f_19 [NUMERICAL]
15 : f_10 [NUMERICAL]
14 : f_9 [NUMERICAL]
13 : f_3 [NUMERICAL]
7 : f_23 [NUMERICAL]
6 : f_24 [NUMERICAL]
5 : f_4 [NUMERICAL]
5 : f_25 [NUMERICAL]
5 : f_21 [NUMERICAL]
5 : f_11 [NUMERICAL]
4 : f_22 [NUMERICAL]
3 : f_20 [NUMERICAL]
3 : f_12 [NUMERICAL]
2 : f_6 [NUMERICAL]
2 : f_16 [NUMERICAL]
1 : f_5 [NUMERICAL]
1 : f_2 [NUMERICAL]
1 : f_18 [NUMERICAL]
1 : f_17 [NUMERICAL]
1 : f_13 [NUMERICAL]
Attribute in nodes with depth <= 0:
4 : f_4 [NUMERICAL]
4 : f_3 [NUMERICAL]
3 : f_8 [NUMERICAL]
1 : f_9 [NUMERICAL]
Attribute in nodes with depth <= 1:
11 : f_9 [NUMERICAL]
9 : f_8 [NUMERICAL]
4 : f_4 [NUMERICAL]
4 : f_3 [NUMERICAL]
1 : f_25 [NUMERICAL]
1 : f_24 [NUMERICAL]
1 : f_23 [NUMERICAL]
1 : f_22 [NUMERICAL]
1 : f_19 [NUMERICAL]
1 : f_11 [NUMERICAL]
Attribute in nodes with depth <= 2:
15 : f_8 [NUMERICAL]
12 : f_9 [NUMERICAL]
11 : f_3 [NUMERICAL]
6 : f_19 [NUMERICAL]
5 : f_4 [NUMERICAL]
2 : f_25 [NUMERICAL]
2 : f_11 [NUMERICAL]
2 : f_10 [NUMERICAL]
1 : f_24 [NUMERICAL]
1 : f_23 [NUMERICAL]
1 : f_22 [NUMERICAL]
1 : f_18 [NUMERICAL]
1 : f_17 [NUMERICAL]
Attribute in nodes with depth <= 3:
22 : f_8 [NUMERICAL]
13 : f_9 [NUMERICAL]
11 : f_3 [NUMERICAL]
10 : f_19 [NUMERICAL]
9 : f_10 [NUMERICAL]
5 : f_4 [NUMERICAL]
5 : f_23 [NUMERICAL]
5 : f_11 [NUMERICAL]
4 : f_25 [NUMERICAL]
4 : f_22 [NUMERICAL]
4 : f_21 [NUMERICAL]
3 : f_24 [NUMERICAL]
2 : f_12 [NUMERICAL]
1 : f_18 [NUMERICAL]
1 : f_17 [NUMERICAL]
Attribute in nodes with depth <= 5:
25 : f_8 [NUMERICAL]
18 : f_19 [NUMERICAL]
15 : f_10 [NUMERICAL]
14 : f_9 [NUMERICAL]
13 : f_3 [NUMERICAL]
7 : f_23 [NUMERICAL]
6 : f_24 [NUMERICAL]
5 : f_4 [NUMERICAL]
5 : f_25 [NUMERICAL]
5 : f_21 [NUMERICAL]
5 : f_11 [NUMERICAL]
4 : f_22 [NUMERICAL]
3 : f_20 [NUMERICAL]
3 : f_12 [NUMERICAL]
2 : f_6 [NUMERICAL]
2 : f_16 [NUMERICAL]
1 : f_5 [NUMERICAL]
1 : f_2 [NUMERICAL]
1 : f_18 [NUMERICAL]
1 : f_17 [NUMERICAL]
1 : f_13 [NUMERICAL]
Condition type in nodes:
137 : HigherCondition
Condition type in nodes with depth <= 0:
12 : HigherCondition
Condition type in nodes with depth <= 1:
34 : HigherCondition
Condition type in nodes with depth <= 2:
60 : HigherCondition
Condition type in nodes with depth <= 3:
99 : HigherCondition
Condition type in nodes with depth <= 5:
137 : HigherCondition
Training logs:
Number of iteration to final model: 12
Iter:1 train-loss:-0.346669 valid-loss:-0.262935 train-NDCG@5:0.346669 valid-NDCG@5:0.262935
Iter:2 train-loss:-0.412635 valid-loss:-0.335301 train-NDCG@5:0.412635 valid-NDCG@5:0.335301
Iter:3 train-loss:-0.468270 valid-loss:-0.341295 train-NDCG@5:0.468270 valid-NDCG@5:0.341295
Iter:4 train-loss:-0.481511 valid-loss:-0.301897 train-NDCG@5:0.481511 valid-NDCG@5:0.301897
Iter:5 train-loss:-0.473165 valid-loss:-0.394670 train-NDCG@5:0.473165 valid-NDCG@5:0.394670
Iter:6 train-loss:-0.496260 valid-loss:-0.415201 train-NDCG@5:0.496260 valid-NDCG@5:0.415201
Iter:16 train-loss:-0.526791 valid-loss:-0.380900 train-NDCG@5:0.526791 valid-NDCG@5:0.380900
Iter:26 train-loss:-0.560398 valid-loss:-0.367496 train-NDCG@5:0.560398 valid-NDCG@5:0.367496
Iter:36 train-loss:-0.584252 valid-loss:-0.341845 train-NDCG@5:0.584252 valid-NDCG@5:0.341845
And if you are curious, you can also plot the model:
tfdf.model_plotter.plot_model_in_colab(model, tree_idx=0, max_depth=3)
Predicting with a ranking model
For an incoming query, we can use our ranking model to predict the relevance of a stack of documents. In practice this means that for each query, we must come up with a set of documents that may or may not be relevant to the query. We call these documents our candidate documents. For each pair query/candidate document, we can compute the same features used during training. This is our serving dataset.
Going back to the example from the beginning of this tutorial, the serving dataset might look like this:
| query | document_id | feature_1 | feature_2 |
|---|---|---|---|
| fish | 32 | 0.3 | blue |
| fish | 33 | 1.0 | green |
| fish | 34 | 0.4 | blue |
| fish | 35 | NA | brown |
Observe that relevance is not part of the serving dataset, since this is what the model is trying to predict.
The serving dataset is fed to the TF-DF model and assigns a relevance score to each document.
| query | document_id | feature_1 | feature_2 | relevance |
|---|---|---|---|---|
| fish | 32 | 0.3 | blue | 0.325 |
| fish | 33 | 1.0 | green | 0.125 |
| fish | 34 | 0.4 | blue | 0.155 |
| fish | 35 | NA | brown | 0.593 |
This means that the document with document_id 35 is predicted to be most relevant for query "fish".
Let's try to do this with our real model.
# Path to a test dataset using libsvm format.
test_dataset_path = os.path.join(os.path.dirname(archive_path),"OHSUMED/Data/Fold1/testset.txt")
# Convert the dataset.
csv_test_dataset_path="/tmp/ohsumed_test.csv"
convert_libsvm_to_csv(raw_dataset_path, csv_test_dataset_path)
# Load a dataset into a Pandas Dataframe.
test_dataset_df = pd.read_csv(csv_test_dataset_path)
# Display the first 3 examples.
test_dataset_df.head(3)
Suppose our query is "g_5" and the test dataset already contains the candidate documents for this query.
# Filter by "g_5"
serving_dataset_df = test_dataset_df[test_dataset_df['group'] == 'g_5']
# Remove the columns for group and relevance, not needed for predictions.
serving_dataset_df = serving_dataset_df.drop(['relevance', 'group'], axis=1)
# Convert to a Tensorflow dataset
serving_dataset_ds = tfdf.keras.pd_dataframe_to_tf_dataset(serving_dataset_df, task=tfdf.keras.Task.RANKING)
# Run predictions with on all candidate documents
predictions = model.predict(serving_dataset_ds)
1/1 [==============================] - 0s 286ms/step
We can use add the predictions to the dataframe and use them to find the documents with the highest scores.
serving_dataset_df['prediction_score'] = predictions
serving_dataset_df.sort_values(by=['prediction_score'], ascending=False).head()
