 View on TensorFlow.org View on TensorFlow.org
|
 Run in Google Colab Run in Google Colab
|
 View on GitHub View on GitHub
|
 Download notebook Download notebook
|
 See TF Hub model See TF Hub model
|
The CORD-19 Swivel text embedding module from TF-Hub (https://tfhub.dev/tensorflow/cord-19/swivel-128d/3) was built to support researchers analyzing natural languages text related to COVID-19. These embeddings were trained on the titles, authors, abstracts, body texts, and reference titles of articles in the CORD-19 dataset.
In this colab we will:
- Analyze semantically similar words in the embedding space
- Train a classifier on the SciCite dataset using the CORD-19 embeddings
Setup
import functools
import itertools
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
import pandas as pd
import tensorflow as tf
import tensorflow_datasets as tfds
import tensorflow_hub as hub
from tqdm import trange
2023-10-09 22:29:39.063949: E tensorflow/compiler/xla/stream_executor/cuda/cuda_dnn.cc:9342] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered 2023-10-09 22:29:39.064004: E tensorflow/compiler/xla/stream_executor/cuda/cuda_fft.cc:609] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered 2023-10-09 22:29:39.064046: E tensorflow/compiler/xla/stream_executor/cuda/cuda_blas.cc:1518] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
Analyze the embeddings
Let's start off by analyzing the embedding by calculating and plotting a correlation matrix between different terms. If the embedding learned to successfully capture the meaning of different words, the embedding vectors of semantically similar words should be close together. Let's take a look at some COVID-19 related terms.
# Use the inner product between two embedding vectors as the similarity measure
def plot_correlation(labels, features):
corr = np.inner(features, features)
corr /= np.max(corr)
sns.heatmap(corr, xticklabels=labels, yticklabels=labels)
# Generate embeddings for some terms
queries = [
# Related viruses
'coronavirus', 'SARS', 'MERS',
# Regions
'Italy', 'Spain', 'Europe',
# Symptoms
'cough', 'fever', 'throat'
]
module = hub.load('https://tfhub.dev/tensorflow/cord-19/swivel-128d/3')
embeddings = module(queries)
plot_correlation(queries, embeddings)
2023-10-09 22:29:44.568345: E tensorflow/compiler/xla/stream_executor/cuda/cuda_driver.cc:268] failed call to cuInit: CUDA_ERROR_NO_DEVICE: no CUDA-capable device is detected
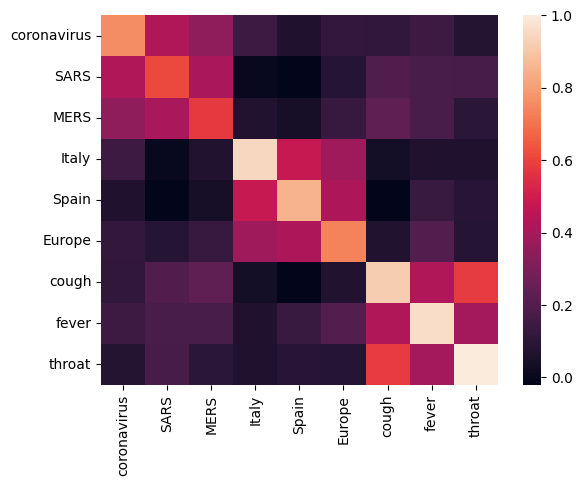
We can see that the embedding successfully captured the meaning of the different terms. Each word is similar to the other words of its cluster (i.e. "coronavirus" highly correlates with "SARS" and "MERS"), while they are different from terms of other clusters (i.e. the similarity between "SARS" and "Spain" is close to 0).
Now let's see how we can use these embeddings to solve a specific task.
SciCite: Citation Intent Classification
This section shows how one can use the embedding for downstream tasks such as text classification. We'll use the SciCite dataset from TensorFlow Datasets to classify citation intents in academic papers. Given a sentence with a citation from an academic paper, classify whether the main intent of the citation is as background information, use of methods, or comparing results.
builder = tfds.builder(name='scicite')
builder.download_and_prepare()
train_data, validation_data, test_data = builder.as_dataset(
split=('train', 'validation', 'test'),
as_supervised=True)
Let's take a look at a few labeled examples from the training set
NUM_EXAMPLES = 10
TEXT_FEATURE_NAME = builder.info.supervised_keys[0]
LABEL_NAME = builder.info.supervised_keys[1]
def label2str(numeric_label):
m = builder.info.features[LABEL_NAME].names
return m[numeric_label]
data = next(iter(train_data.batch(NUM_EXAMPLES)))
pd.DataFrame({
TEXT_FEATURE_NAME: [ex.numpy().decode('utf8') for ex in data[0]],
LABEL_NAME: [label2str(x) for x in data[1]]
})
Training a citaton intent classifier
We'll train a classifier on the SciCite dataset using Keras. Let's build a model which use the CORD-19 embeddings with a classification layer on top.
Hyperparameters
EMBEDDING = 'https://tfhub.dev/tensorflow/cord-19/swivel-128d/3'
TRAINABLE_MODULE = False
hub_layer = hub.KerasLayer(EMBEDDING, input_shape=[],
dtype=tf.string, trainable=TRAINABLE_MODULE)
model = tf.keras.Sequential()
model.add(hub_layer)
model.add(tf.keras.layers.Dense(3))
model.summary()
model.compile(optimizer='adam',
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=['accuracy'])
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
keras_layer (KerasLayer) (None, 128) 17301632
dense (Dense) (None, 3) 387
=================================================================
Total params: 17302019 (132.00 MB)
Trainable params: 387 (1.51 KB)
Non-trainable params: 17301632 (132.00 MB)
_________________________________________________________________
Train and evaluate the model
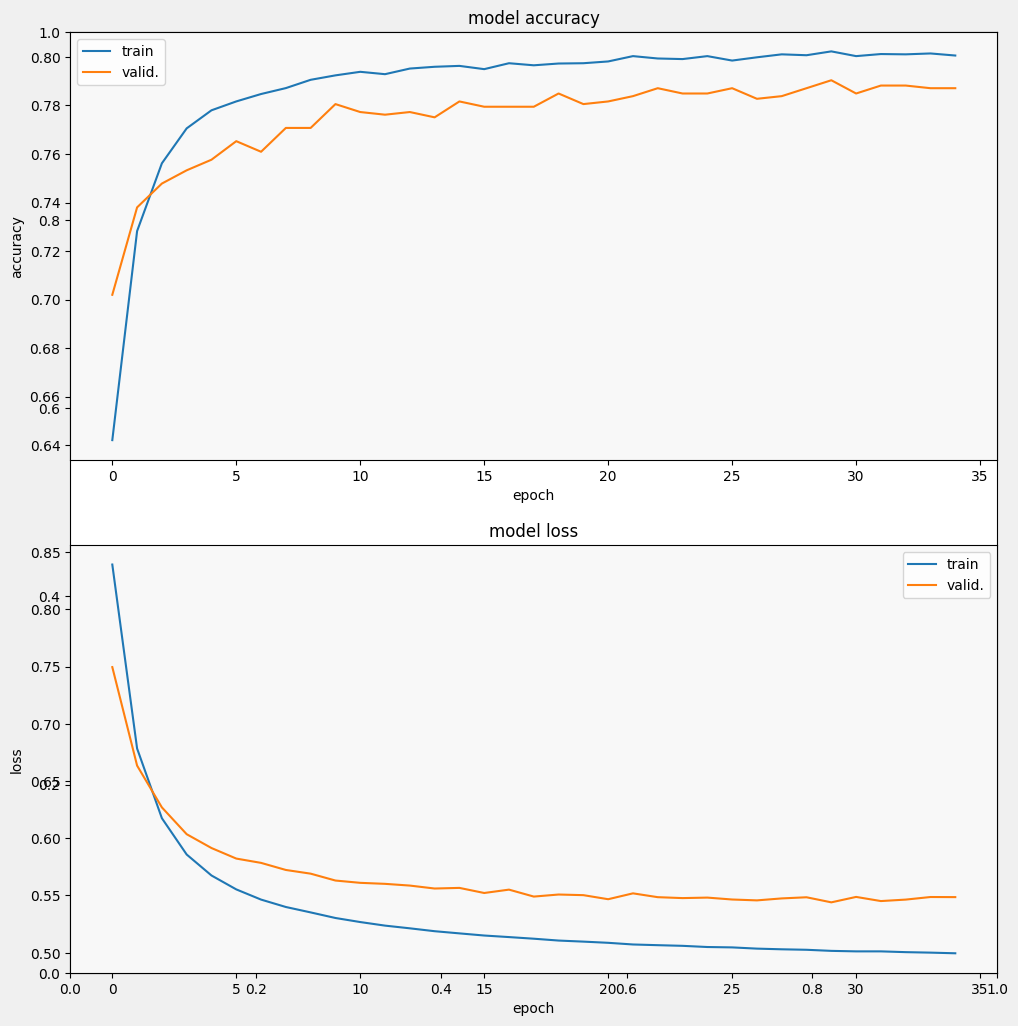
Let's train and evaluate the model to see the performance on the SciCite task
EPOCHS = 35
BATCH_SIZE = 32
history = model.fit(train_data.shuffle(10000).batch(BATCH_SIZE),
epochs=EPOCHS,
validation_data=validation_data.batch(BATCH_SIZE),
verbose=1)
Epoch 1/35 257/257 [==============================] - 2s 4ms/step - loss: 0.8391 - accuracy: 0.6421 - val_loss: 0.7495 - val_accuracy: 0.7020 Epoch 2/35 257/257 [==============================] - 1s 3ms/step - loss: 0.6784 - accuracy: 0.7282 - val_loss: 0.6634 - val_accuracy: 0.7380 Epoch 3/35 257/257 [==============================] - 1s 3ms/step - loss: 0.6175 - accuracy: 0.7562 - val_loss: 0.6269 - val_accuracy: 0.7478 Epoch 4/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5858 - accuracy: 0.7706 - val_loss: 0.6035 - val_accuracy: 0.7533 Epoch 5/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5674 - accuracy: 0.7780 - val_loss: 0.5914 - val_accuracy: 0.7576 Epoch 6/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5553 - accuracy: 0.7817 - val_loss: 0.5822 - val_accuracy: 0.7653 Epoch 7/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5464 - accuracy: 0.7847 - val_loss: 0.5784 - val_accuracy: 0.7609 Epoch 8/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5399 - accuracy: 0.7872 - val_loss: 0.5723 - val_accuracy: 0.7707 Epoch 9/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5352 - accuracy: 0.7906 - val_loss: 0.5690 - val_accuracy: 0.7707 Epoch 10/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5303 - accuracy: 0.7924 - val_loss: 0.5630 - val_accuracy: 0.7806 Epoch 11/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5268 - accuracy: 0.7939 - val_loss: 0.5610 - val_accuracy: 0.7773 Epoch 12/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5236 - accuracy: 0.7929 - val_loss: 0.5601 - val_accuracy: 0.7762 Epoch 13/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5213 - accuracy: 0.7952 - val_loss: 0.5586 - val_accuracy: 0.7773 Epoch 14/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5188 - accuracy: 0.7959 - val_loss: 0.5560 - val_accuracy: 0.7751 Epoch 15/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5169 - accuracy: 0.7963 - val_loss: 0.5566 - val_accuracy: 0.7817 Epoch 16/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5150 - accuracy: 0.7950 - val_loss: 0.5521 - val_accuracy: 0.7795 Epoch 17/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5136 - accuracy: 0.7974 - val_loss: 0.5551 - val_accuracy: 0.7795 Epoch 18/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5122 - accuracy: 0.7966 - val_loss: 0.5490 - val_accuracy: 0.7795 Epoch 19/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5106 - accuracy: 0.7973 - val_loss: 0.5508 - val_accuracy: 0.7849 Epoch 20/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5097 - accuracy: 0.7974 - val_loss: 0.5503 - val_accuracy: 0.7806 Epoch 21/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5086 - accuracy: 0.7981 - val_loss: 0.5467 - val_accuracy: 0.7817 Epoch 22/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5072 - accuracy: 0.8003 - val_loss: 0.5518 - val_accuracy: 0.7838 Epoch 23/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5066 - accuracy: 0.7994 - val_loss: 0.5485 - val_accuracy: 0.7871 Epoch 24/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5060 - accuracy: 0.7991 - val_loss: 0.5477 - val_accuracy: 0.7849 Epoch 25/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5049 - accuracy: 0.8003 - val_loss: 0.5481 - val_accuracy: 0.7849 Epoch 26/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5046 - accuracy: 0.7985 - val_loss: 0.5465 - val_accuracy: 0.7871 Epoch 27/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5035 - accuracy: 0.7999 - val_loss: 0.5457 - val_accuracy: 0.7828 Epoch 28/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5030 - accuracy: 0.8011 - val_loss: 0.5474 - val_accuracy: 0.7838 Epoch 29/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5025 - accuracy: 0.8007 - val_loss: 0.5484 - val_accuracy: 0.7871 Epoch 30/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5016 - accuracy: 0.8023 - val_loss: 0.5440 - val_accuracy: 0.7904 Epoch 31/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5011 - accuracy: 0.8003 - val_loss: 0.5487 - val_accuracy: 0.7849 Epoch 32/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5011 - accuracy: 0.8012 - val_loss: 0.5451 - val_accuracy: 0.7882 Epoch 33/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5005 - accuracy: 0.8011 - val_loss: 0.5464 - val_accuracy: 0.7882 Epoch 34/35 257/257 [==============================] - 1s 3ms/step - loss: 0.5000 - accuracy: 0.8014 - val_loss: 0.5486 - val_accuracy: 0.7871 Epoch 35/35 257/257 [==============================] - 1s 3ms/step - loss: 0.4995 - accuracy: 0.8006 - val_loss: 0.5485 - val_accuracy: 0.7871
from matplotlib import pyplot as plt
def display_training_curves(training, validation, title, subplot):
if subplot%10==1: # set up the subplots on the first call
plt.subplots(figsize=(10,10), facecolor='#F0F0F0')
plt.tight_layout()
ax = plt.subplot(subplot)
ax.set_facecolor('#F8F8F8')
ax.plot(training)
ax.plot(validation)
ax.set_title('model '+ title)
ax.set_ylabel(title)
ax.set_xlabel('epoch')
ax.legend(['train', 'valid.'])
display_training_curves(history.history['accuracy'], history.history['val_accuracy'], 'accuracy', 211)
display_training_curves(history.history['loss'], history.history['val_loss'], 'loss', 212)
Evaluate the model
And let's see how the model performs. Two values will be returned. Loss (a number which represents our error, lower values are better), and accuracy.
results = model.evaluate(test_data.batch(512), verbose=2)
for name, value in zip(model.metrics_names, results):
print('%s: %.3f' % (name, value))
4/4 - 0s - loss: 0.5406 - accuracy: 0.7805 - 293ms/epoch - 73ms/step loss: 0.541 accuracy: 0.781
We can see that the loss quickly decreases while especially the accuracy rapidly increases. Let's plot some examples to check how the prediction relates to the true labels:
prediction_dataset = next(iter(test_data.batch(20)))
prediction_texts = [ex.numpy().decode('utf8') for ex in prediction_dataset[0]]
prediction_labels = [label2str(x) for x in prediction_dataset[1]]
predictions = [
label2str(x) for x in np.argmax(model.predict(prediction_texts), axis=-1)]
pd.DataFrame({
TEXT_FEATURE_NAME: prediction_texts,
LABEL_NAME: prediction_labels,
'prediction': predictions
})
1/1 [==============================] - 0s 122ms/step
We can see that for this random sample, the model predicts the correct label most of the times, indicating that it can embed scientific sentences pretty well.
What's next?
Now that you've gotten to know a bit more about the CORD-19 Swivel embeddings from TF-Hub, we encourage you to participate in the CORD-19 Kaggle competition to contribute to gaining scientific insights from COVID-19 related academic texts.
- Participate in the CORD-19 Kaggle Challenge
- Learn more about the COVID-19 Open Research Dataset (CORD-19)
- See documentation and more about the TF-Hub embeddings at https://tfhub.dev/tensorflow/cord-19/swivel-128d/3
- Explore the CORD-19 embedding space with the TensorFlow Embedding Projector
