 Voir sur TensorFlow.org Voir sur TensorFlow.org |  Exécuter dans Google Colab Exécuter dans Google Colab |  Voir sur GitHub Voir sur GitHub |  Télécharger le cahier Télécharger le cahier |  Voir le modèle TF Hub Voir le modèle TF Hub |
Le CORD-19 Module intégration texte pivotant de TF-Hub ( https://tfhub.dev/tensorflow/cord-19/swivel-128d/3 ) a été conçu pour appuyer les chercheurs qui analysent le texte des langues naturelles liées à Covid-19. Ces incorporations ont été formés sur les titres, auteurs, résumés, textes du corps, et les titres de référence des articles dans le jeu de données CORD-19 .
Dans cette collaboration, nous allons :
- Analyser les mots sémantiquement similaires dans l'espace d'inclusion
- Former un classificateur sur l'ensemble de données SciCite à l'aide des plongements CORD-19
Installer
import functools
import itertools
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
import pandas as pd
import tensorflow as tf
import tensorflow_datasets as tfds
import tensorflow_hub as hub
from tqdm import trange
Analyser les encastrements
Commençons par analyser le plongement en calculant et en traçant une matrice de corrélation entre différents termes. Si l'intégration a appris à capturer avec succès le sens de différents mots, les vecteurs d'intégration de mots sémantiquement similaires doivent être proches les uns des autres. Jetons un coup d'œil à quelques termes liés à COVID-19.
# Use the inner product between two embedding vectors as the similarity measure
def plot_correlation(labels, features):
corr = np.inner(features, features)
corr /= np.max(corr)
sns.heatmap(corr, xticklabels=labels, yticklabels=labels)
# Generate embeddings for some terms
queries = [
# Related viruses
'coronavirus', 'SARS', 'MERS',
# Regions
'Italy', 'Spain', 'Europe',
# Symptoms
'cough', 'fever', 'throat'
]
module = hub.load('https://tfhub.dev/tensorflow/cord-19/swivel-128d/3')
embeddings = module(queries)
plot_correlation(queries, embeddings)

Nous pouvons voir que l'intégration a réussi à capturer le sens des différents termes. Chaque mot est similaire aux autres mots de son groupe (c. proche de 0).
Voyons maintenant comment nous pouvons utiliser ces intégrations pour résoudre une tâche spécifique.
SciCite : classification des intentions de citation
Cette section montre comment utiliser l'intégration pour des tâches en aval telles que la classification de texte. Nous allons utiliser l' ensemble de données SciCite de tensorflow datasets pour classer les intentions de citation dans les journaux académiques. À partir d'une phrase avec une citation d'un article universitaire, indiquez si l'intention principale de la citation est une information de base, l'utilisation de méthodes ou la comparaison de résultats.
builder = tfds.builder(name='scicite')
builder.download_and_prepare()
train_data, validation_data, test_data = builder.as_dataset(
split=('train', 'validation', 'test'),
as_supervised=True)
Jetons un coup d'œil à quelques exemples étiquetés de l'ensemble de formation
NUM_EXAMPLES = 10
TEXT_FEATURE_NAME = builder.info.supervised_keys[0]
LABEL_NAME = builder.info.supervised_keys[1]
def label2str(numeric_label):
m = builder.info.features[LABEL_NAME].names
return m[numeric_label]
data = next(iter(train_data.batch(NUM_EXAMPLES)))
pd.DataFrame({
TEXT_FEATURE_NAME: [ex.numpy().decode('utf8') for ex in data[0]],
LABEL_NAME: [label2str(x) for x in data[1]]
})
Formation d'un classificateur d'intention citaton
Nous allons former un classificateur sur l' ensemble de données SciCite en utilisant Keras. Construisons un modèle qui utilise les plongements CORD-19 avec une couche de classification au-dessus.
Hyperparamètres
EMBEDDING = 'https://tfhub.dev/tensorflow/cord-19/swivel-128d/3'
TRAINABLE_MODULE = False
hub_layer = hub.KerasLayer(EMBEDDING, input_shape=[],
dtype=tf.string, trainable=TRAINABLE_MODULE)
model = tf.keras.Sequential()
model.add(hub_layer)
model.add(tf.keras.layers.Dense(3))
model.summary()
model.compile(optimizer='adam',
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=['accuracy'])
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
keras_layer (KerasLayer) (None, 128) 17301632
dense (Dense) (None, 3) 387
=================================================================
Total params: 17,302,019
Trainable params: 387
Non-trainable params: 17,301,632
_________________________________________________________________
Former et évaluer le modèle
Entraînons et évaluons le modèle pour voir les performances sur la tâche SciCite
EPOCHS = 35
BATCH_SIZE = 32
history = model.fit(train_data.shuffle(10000).batch(BATCH_SIZE),
epochs=EPOCHS,
validation_data=validation_data.batch(BATCH_SIZE),
verbose=1)
Epoch 1/35 257/257 [==============================] - 3s 7ms/step - loss: 0.9244 - accuracy: 0.5924 - val_loss: 0.7915 - val_accuracy: 0.6627 Epoch 2/35 257/257 [==============================] - 2s 5ms/step - loss: 0.7097 - accuracy: 0.7152 - val_loss: 0.6799 - val_accuracy: 0.7358 Epoch 3/35 257/257 [==============================] - 2s 7ms/step - loss: 0.6317 - accuracy: 0.7551 - val_loss: 0.6285 - val_accuracy: 0.7544 Epoch 4/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5938 - accuracy: 0.7687 - val_loss: 0.6032 - val_accuracy: 0.7566 Epoch 5/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5724 - accuracy: 0.7750 - val_loss: 0.5871 - val_accuracy: 0.7653 Epoch 6/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5580 - accuracy: 0.7825 - val_loss: 0.5800 - val_accuracy: 0.7653 Epoch 7/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5484 - accuracy: 0.7870 - val_loss: 0.5711 - val_accuracy: 0.7718 Epoch 8/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5417 - accuracy: 0.7896 - val_loss: 0.5648 - val_accuracy: 0.7806 Epoch 9/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5356 - accuracy: 0.7902 - val_loss: 0.5628 - val_accuracy: 0.7740 Epoch 10/35 257/257 [==============================] - 2s 7ms/step - loss: 0.5313 - accuracy: 0.7903 - val_loss: 0.5581 - val_accuracy: 0.7849 Epoch 11/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5277 - accuracy: 0.7928 - val_loss: 0.5555 - val_accuracy: 0.7838 Epoch 12/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5242 - accuracy: 0.7940 - val_loss: 0.5528 - val_accuracy: 0.7849 Epoch 13/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5215 - accuracy: 0.7947 - val_loss: 0.5522 - val_accuracy: 0.7828 Epoch 14/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5190 - accuracy: 0.7961 - val_loss: 0.5527 - val_accuracy: 0.7751 Epoch 15/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5176 - accuracy: 0.7940 - val_loss: 0.5492 - val_accuracy: 0.7806 Epoch 16/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5154 - accuracy: 0.7978 - val_loss: 0.5500 - val_accuracy: 0.7817 Epoch 17/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5136 - accuracy: 0.7968 - val_loss: 0.5488 - val_accuracy: 0.7795 Epoch 18/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5127 - accuracy: 0.7967 - val_loss: 0.5504 - val_accuracy: 0.7838 Epoch 19/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5111 - accuracy: 0.7970 - val_loss: 0.5470 - val_accuracy: 0.7860 Epoch 20/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5101 - accuracy: 0.7972 - val_loss: 0.5471 - val_accuracy: 0.7871 Epoch 21/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5082 - accuracy: 0.7997 - val_loss: 0.5483 - val_accuracy: 0.7784 Epoch 22/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5077 - accuracy: 0.7995 - val_loss: 0.5471 - val_accuracy: 0.7860 Epoch 23/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5064 - accuracy: 0.8012 - val_loss: 0.5439 - val_accuracy: 0.7871 Epoch 24/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5057 - accuracy: 0.7990 - val_loss: 0.5476 - val_accuracy: 0.7882 Epoch 25/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5050 - accuracy: 0.7996 - val_loss: 0.5442 - val_accuracy: 0.7937 Epoch 26/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5045 - accuracy: 0.7999 - val_loss: 0.5455 - val_accuracy: 0.7860 Epoch 27/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5032 - accuracy: 0.7991 - val_loss: 0.5435 - val_accuracy: 0.7893 Epoch 28/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5034 - accuracy: 0.8022 - val_loss: 0.5431 - val_accuracy: 0.7882 Epoch 29/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5025 - accuracy: 0.8017 - val_loss: 0.5441 - val_accuracy: 0.7937 Epoch 30/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5017 - accuracy: 0.8013 - val_loss: 0.5463 - val_accuracy: 0.7838 Epoch 31/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5015 - accuracy: 0.8017 - val_loss: 0.5453 - val_accuracy: 0.7871 Epoch 32/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5011 - accuracy: 0.8014 - val_loss: 0.5448 - val_accuracy: 0.7915 Epoch 33/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5006 - accuracy: 0.8025 - val_loss: 0.5432 - val_accuracy: 0.7893 Epoch 34/35 257/257 [==============================] - 2s 5ms/step - loss: 0.5005 - accuracy: 0.8008 - val_loss: 0.5448 - val_accuracy: 0.7904 Epoch 35/35 257/257 [==============================] - 2s 5ms/step - loss: 0.4996 - accuracy: 0.8016 - val_loss: 0.5448 - val_accuracy: 0.7915
from matplotlib import pyplot as plt
def display_training_curves(training, validation, title, subplot):
if subplot%10==1: # set up the subplots on the first call
plt.subplots(figsize=(10,10), facecolor='#F0F0F0')
plt.tight_layout()
ax = plt.subplot(subplot)
ax.set_facecolor('#F8F8F8')
ax.plot(training)
ax.plot(validation)
ax.set_title('model '+ title)
ax.set_ylabel(title)
ax.set_xlabel('epoch')
ax.legend(['train', 'valid.'])
display_training_curves(history.history['accuracy'], history.history['val_accuracy'], 'accuracy', 211)
display_training_curves(history.history['loss'], history.history['val_loss'], 'loss', 212)
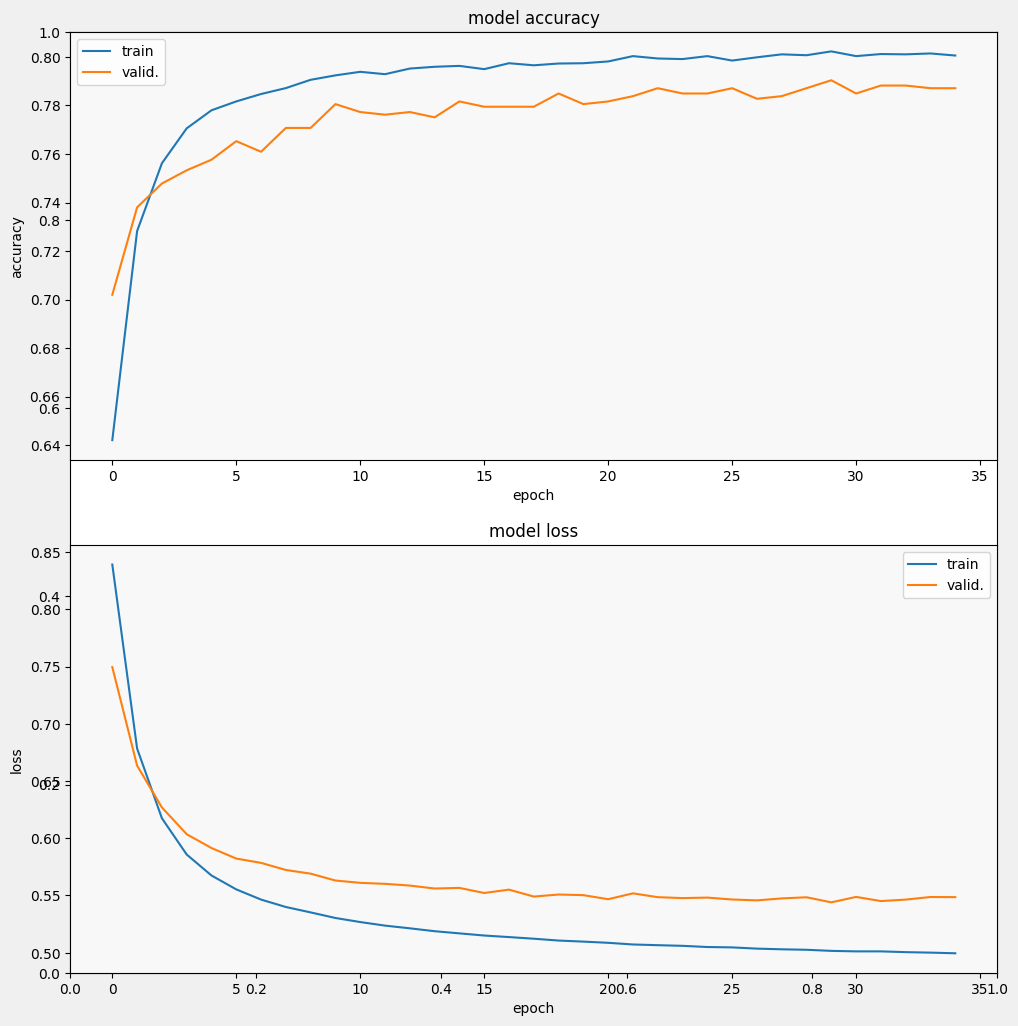
Évaluer le modèle
Et voyons comment le modèle fonctionne. Deux valeurs seront renvoyées. Perte (un nombre qui représente notre erreur, les valeurs inférieures sont meilleures) et précision.
results = model.evaluate(test_data.batch(512), verbose=2)
for name, value in zip(model.metrics_names, results):
print('%s: %.3f' % (name, value))
4/4 - 0s - loss: 0.5357 - accuracy: 0.7891 - 441ms/epoch - 110ms/step loss: 0.536 accuracy: 0.789
On peut voir que la perte diminue rapidement tandis que surtout la précision augmente rapidement. Traçons quelques exemples pour vérifier comment la prédiction se rapporte aux vraies étiquettes :
prediction_dataset = next(iter(test_data.batch(20)))
prediction_texts = [ex.numpy().decode('utf8') for ex in prediction_dataset[0]]
prediction_labels = [label2str(x) for x in prediction_dataset[1]]
predictions = [
label2str(x) for x in np.argmax(model.predict(prediction_texts), axis=-1)]
pd.DataFrame({
TEXT_FEATURE_NAME: prediction_texts,
LABEL_NAME: prediction_labels,
'prediction': predictions
})
Nous pouvons voir que pour cet échantillon aléatoire, le modèle prédit la bonne étiquette la plupart du temps, indiquant qu'il peut assez bien intégrer des phrases scientifiques.
Et après?
Maintenant que vous en savez un peu plus sur les encastrements CORD-19 Swivel de TF-Hub, nous vous encourageons à participer au concours CORD-19 Kaggle pour contribuer à acquérir des connaissances scientifiques à partir de textes académiques liés à COVID-19.
- Participer à la CORD-19 Kaggle Défi
- En savoir plus sur le Covid-19 Recherche ouverte Dataset (CORD-19)
- Voir la documentation et plus sur les incorporations TF-Hub à https://tfhub.dev/tensorflow/cord-19/swivel-128d/3
- Explorez l'espace plongement CORD-19 avec le tensorflow Embedding projecteur
