 TensorFlow.org'da görüntüleyin TensorFlow.org'da görüntüleyin |  Google Colab'da çalıştırın Google Colab'da çalıştırın |  Kaynağı GitHub'da görüntüleyin Kaynağı GitHub'da görüntüleyin |  Not defterini indir Not defterini indir |
Bu İşbirliğinde, TensorFlow Olasılığının bazı temel özelliklerini keşfediyoruz.
Bağımlılıklar ve Ön Koşullar
İçe aktarmak
from pprint import pprint
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
import tensorflow.compat.v2 as tf
tf.enable_v2_behavior()
import tensorflow_probability as tfp
sns.reset_defaults()
sns.set_context(context='talk',font_scale=0.7)
plt.rcParams['image.cmap'] = 'viridis'
%matplotlib inline
tfd = tfp.distributions
tfb = tfp.bijectors
araçlar
def print_subclasses_from_module(module, base_class, maxwidth=80):
import functools, inspect, sys
subclasses = [name for name, obj in inspect.getmembers(module)
if inspect.isclass(obj) and issubclass(obj, base_class)]
def red(acc, x):
if not acc or len(acc[-1]) + len(x) + 2 > maxwidth:
acc.append(x)
else:
acc[-1] += ", " + x
return acc
print('\n'.join(functools.reduce(red, subclasses, [])))
anahat
- TensorFlow
- TensorFlow Olasılığı
- dağıtımlar
- Bijektörler
- MCMC
- ...ve dahası!
Önsöz: TensorFlow
TensorFlow, bilimsel bir bilgi işlem kitaplığıdır.
Destekler
- çok sayıda matematiksel işlem
- verimli vektörleştirilmiş hesaplama
- kolay donanım hızlandırma
- otomatik farklılaşma
vektörleştirme
- Vektörleştirme işleri hızlandırır!
- Aynı zamanda şekiller hakkında çok düşündüğümüz anlamına gelir.
mats = tf.random.uniform(shape=[1000, 10, 10])
vecs = tf.random.uniform(shape=[1000, 10, 1])
def for_loop_solve():
return np.array(
[tf.linalg.solve(mats[i, ...], vecs[i, ...]) for i in range(1000)])
def vectorized_solve():
return tf.linalg.solve(mats, vecs)
# Vectorization for the win!
%timeit for_loop_solve()
%timeit vectorized_solve()
1 loops, best of 3: 2 s per loop 1000 loops, best of 3: 653 µs per loop
Donanım ivmesi
# Code can run seamlessly on a GPU, just change Colab runtime type
# in the 'Runtime' menu.
if tf.test.gpu_device_name() == '/device:GPU:0':
print("Using a GPU")
else:
print("Using a CPU")
Using a CPU
Otomatik Farklılaşma
a = tf.constant(np.pi)
b = tf.constant(np.e)
with tf.GradientTape() as tape:
tape.watch([a, b])
c = .5 * (a**2 + b**2)
grads = tape.gradient(c, [a, b])
print(grads[0])
print(grads[1])
tf.Tensor(3.1415927, shape=(), dtype=float32) tf.Tensor(2.7182817, shape=(), dtype=float32)
TensorFlow Olasılığı
TensorFlow Probability, TensorFlow'da olasılıksal akıl yürütme ve istatistiksel analiz için bir kitaplıktır.
Biz düşük seviyeli modüler bileşenlerin bileşimi sayesinde modelleme, çıkarım ve eleştiri destekler.
Düşük seviyeli yapı taşları
- dağıtımlar
- Bijektörler
Yüksek(er)-seviye yapılar
- Markov zinciri Monte Carlo
- Olasılık Katmanları
- Yapısal Zaman Serileri
- Genelleştirilmiş Doğrusal Modeller
- Optimize Ediciler
dağıtımlar
Bir tfp.distributions.Distribution : iki temel yöntem ile bir sınıftır sample ve log_prob .
TFP'nin birçok dağıtımı var!
print_subclasses_from_module(tfp.distributions, tfp.distributions.Distribution)
Autoregressive, BatchReshape, Bates, Bernoulli, Beta, BetaBinomial, Binomial Blockwise, Categorical, Cauchy, Chi, Chi2, CholeskyLKJ, ContinuousBernoulli Deterministic, Dirichlet, DirichletMultinomial, Distribution, DoublesidedMaxwell Empirical, ExpGamma, ExpRelaxedOneHotCategorical, Exponential, FiniteDiscrete Gamma, GammaGamma, GaussianProcess, GaussianProcessRegressionModel GeneralizedNormal, GeneralizedPareto, Geometric, Gumbel, HalfCauchy, HalfNormal HalfStudentT, HiddenMarkovModel, Horseshoe, Independent, InverseGamma InverseGaussian, JohnsonSU, JointDistribution, JointDistributionCoroutine JointDistributionCoroutineAutoBatched, JointDistributionNamed JointDistributionNamedAutoBatched, JointDistributionSequential JointDistributionSequentialAutoBatched, Kumaraswamy, LKJ, Laplace LinearGaussianStateSpaceModel, LogLogistic, LogNormal, Logistic, LogitNormal Mixture, MixtureSameFamily, Moyal, Multinomial, MultivariateNormalDiag MultivariateNormalDiagPlusLowRank, MultivariateNormalFullCovariance MultivariateNormalLinearOperator, MultivariateNormalTriL MultivariateStudentTLinearOperator, NegativeBinomial, Normal, OneHotCategorical OrderedLogistic, PERT, Pareto, PixelCNN, PlackettLuce, Poisson PoissonLogNormalQuadratureCompound, PowerSpherical, ProbitBernoulli QuantizedDistribution, RelaxedBernoulli, RelaxedOneHotCategorical, Sample SinhArcsinh, SphericalUniform, StudentT, StudentTProcess TransformedDistribution, Triangular, TruncatedCauchy, TruncatedNormal, Uniform VariationalGaussianProcess, VectorDeterministic, VonMises VonMisesFisher, Weibull, WishartLinearOperator, WishartTriL, Zipf
Basit bir skaler değişkenli Distribution
# A standard normal
normal = tfd.Normal(loc=0., scale=1.)
print(normal)
tfp.distributions.Normal("Normal", batch_shape=[], event_shape=[], dtype=float32)
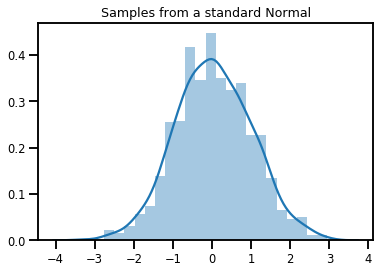
# Plot 1000 samples from a standard normal
samples = normal.sample(1000)
sns.distplot(samples)
plt.title("Samples from a standard Normal")
plt.show()
# Compute the log_prob of a point in the event space of `normal`
normal.log_prob(0.)
<tf.Tensor: shape=(), dtype=float32, numpy=-0.9189385>
# Compute the log_prob of a few points
normal.log_prob([-1., 0., 1.])
<tf.Tensor: shape=(3,), dtype=float32, numpy=array([-1.4189385, -0.9189385, -1.4189385], dtype=float32)>
Dağılımlar ve Şekiller
Numpy ndarrays ve TensorFlow Tensors şekilleri vardır.
TensorFlow Olasılık Distributions belleğin aynı öbek (olsa bile, semantik olarak ayrı parçalar halinde biz bölme şekilleri - şekil anlambilgisi Tensor / ndarray ) Bütün her şey için kullanılır.
- Toplu şekli bir koleksiyon belirtmektedir
Distributionfarklı parametrelerle s - Olay şekli örnekler şeklini gösterir
Distribution.
Parti şekillerini her zaman "sola" ve olay şekillerini "sağ"a koyarız.
Skaler değişkenli bir toplu Distributions
Gruplar "vektörlü" dağılımlar gibidir: hesaplamaları paralel olarak gerçekleşen bağımsız örnekler.
# Create a batch of 3 normals, and plot 1000 samples from each
normals = tfd.Normal([-2.5, 0., 2.5], 1.) # The scale parameter broadacasts!
print("Batch shape:", normals.batch_shape)
print("Event shape:", normals.event_shape)
Batch shape: (3,) Event shape: ()
# Samples' shapes go on the left!
samples = normals.sample(1000)
print("Shape of samples:", samples.shape)
Shape of samples: (1000, 3)
# Sample shapes can themselves be more complicated
print("Shape of samples:", normals.sample([10, 10, 10]).shape)
Shape of samples: (10, 10, 10, 3)
# A batch of normals gives a batch of log_probs.
print(normals.log_prob([-2.5, 0., 2.5]))
tf.Tensor([-0.9189385 -0.9189385 -0.9189385], shape=(3,), dtype=float32)
# The computation broadcasts, so a batch of normals applied to a scalar
# also gives a batch of log_probs.
print(normals.log_prob(0.))
tf.Tensor([-4.0439386 -0.9189385 -4.0439386], shape=(3,), dtype=float32)
# Normal numpy-like broadcasting rules apply!
xs = np.linspace(-6, 6, 200)
try:
normals.log_prob(xs)
except Exception as e:
print("TFP error:", e.message)
TFP error: Incompatible shapes: [200] vs. [3] [Op:SquaredDifference]
# That fails for the same reason this does:
try:
np.zeros(200) + np.zeros(3)
except Exception as e:
print("Numpy error:", e)
Numpy error: operands could not be broadcast together with shapes (200,) (3,)
# But this would work:
a = np.zeros([200, 1]) + np.zeros(3)
print("Broadcast shape:", a.shape)
Broadcast shape: (200, 3)
# And so will this!
xs = np.linspace(-6, 6, 200)[..., np.newaxis]
# => shape = [200, 1]
lps = normals.log_prob(xs)
print("Broadcast log_prob shape:", lps.shape)
Broadcast log_prob shape: (200, 3)
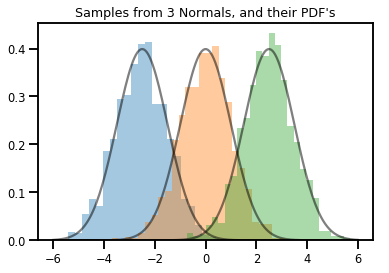
# Summarizing visually
for i in range(3):
sns.distplot(samples[:, i], kde=False, norm_hist=True)
plt.plot(np.tile(xs, 3), normals.prob(xs), c='k', alpha=.5)
plt.title("Samples from 3 Normals, and their PDF's")
plt.show()
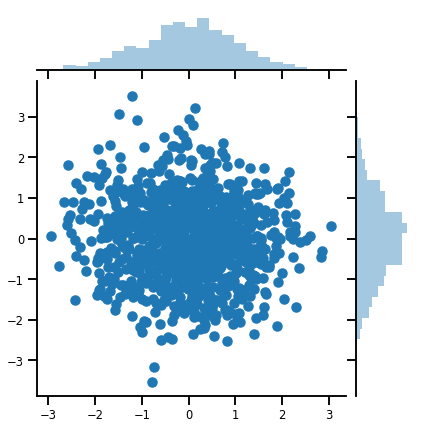
Bir vektör-değişken Distribution
mvn = tfd.MultivariateNormalDiag(loc=[0., 0.], scale_diag = [1., 1.])
print("Batch shape:", mvn.batch_shape)
print("Event shape:", mvn.event_shape)
Batch shape: () Event shape: (2,)
samples = mvn.sample(1000)
print("Samples shape:", samples.shape)
Samples shape: (1000, 2)
g = sns.jointplot(samples[:, 0], samples[:, 1], kind='scatter')
plt.show()
Bir matris-değişken Distribution
lkj = tfd.LKJ(dimension=10, concentration=[1.5, 3.0])
print("Batch shape: ", lkj.batch_shape)
print("Event shape: ", lkj.event_shape)
Batch shape: (2,) Event shape: (10, 10)
samples = lkj.sample()
print("Samples shape: ", samples.shape)
Samples shape: (2, 10, 10)
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(6, 3))
sns.heatmap(samples[0, ...], ax=axes[0], cbar=False)
sns.heatmap(samples[1, ...], ax=axes[1], cbar=False)
fig.tight_layout()
plt.show()
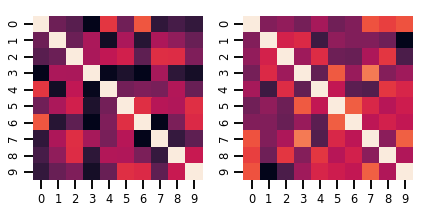
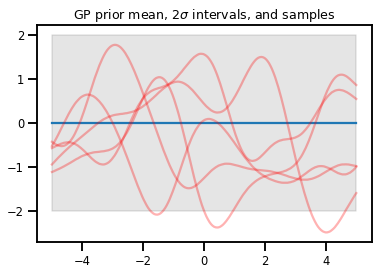
Gauss Süreçleri
kernel = tfp.math.psd_kernels.ExponentiatedQuadratic()
xs = np.linspace(-5., 5., 200).reshape([-1, 1])
gp = tfd.GaussianProcess(kernel, index_points=xs)
print("Batch shape:", gp.batch_shape)
print("Event shape:", gp.event_shape)
Batch shape: () Event shape: (200,)
upper, lower = gp.mean() + [2 * gp.stddev(), -2 * gp.stddev()]
plt.plot(xs, gp.mean())
plt.fill_between(xs[..., 0], upper, lower, color='k', alpha=.1)
for _ in range(5):
plt.plot(xs, gp.sample(), c='r', alpha=.3)
plt.title(r"GP prior mean, $2\sigma$ intervals, and samples")
plt.show()
# *** Bonus question ***
# Why do so many of these functions lie outside the 95% intervals?
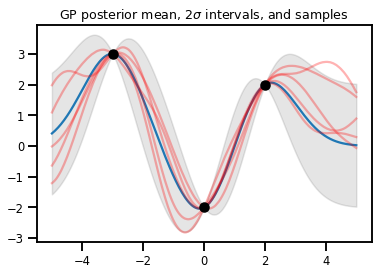
GP Regresyonu
# Suppose we have some observed data
obs_x = [[-3.], [0.], [2.]] # Shape 3x1 (3 1-D vectors)
obs_y = [3., -2., 2.] # Shape 3 (3 scalars)
gprm = tfd.GaussianProcessRegressionModel(kernel, xs, obs_x, obs_y)
upper, lower = gprm.mean() + [2 * gprm.stddev(), -2 * gprm.stddev()]
plt.plot(xs, gprm.mean())
plt.fill_between(xs[..., 0], upper, lower, color='k', alpha=.1)
for _ in range(5):
plt.plot(xs, gprm.sample(), c='r', alpha=.3)
plt.scatter(obs_x, obs_y, c='k', zorder=3)
plt.title(r"GP posterior mean, $2\sigma$ intervals, and samples")
plt.show()
Bijektörler
Bijektörler (çoğunlukla) ters çevrilebilir, pürüzsüz işlevleri temsil eder. Örnek alma ve log_probs hesaplama yeteneğini koruyarak dağılımları dönüştürmek için kullanılabilirler. Bunlar olabilir tfp.bijectors modülü.
Her bijektör en az 3 yöntem uygular:
-
forward, -
inverseve - ve (en azından) bir
forward_log_det_jacobianveinverse_log_det_jacobian.
Bu bileşenlerle, bir dağılımı dönüştürebilir ve yine de sonuçtan örnekler ve log probları alabiliriz!
Matematikte, biraz özensiz
- \(X\) pdf rastgele değişken \(p(x)\)
- \(g\) uzayında bir düz, ters çevrilebilir bir fonksiyondur \(X\)'ler
- \(Y = g(X)\) yeni bir dönüştürülmüş rastgele değişken
- \(p(Y=y) = p(X=g^{-1}(y)) \cdot |\nabla g^{-1}(y)|\)
Önbelleğe almak
Bijektörler ayrıca ileri ve ters hesaplamaları ve log-det-Jacobians'ı önbelleğe alır, bu da potansiyel olarak çok pahalı işlemleri tekrarlamaktan tasarruf etmemizi sağlar!
print_subclasses_from_module(tfp.bijectors, tfp.bijectors.Bijector)
AbsoluteValue, Affine, AffineLinearOperator, AffineScalar, BatchNormalization Bijector, Blockwise, Chain, CholeskyOuterProduct, CholeskyToInvCholesky CorrelationCholesky, Cumsum, DiscreteCosineTransform, Exp, Expm1, FFJORD FillScaleTriL, FillTriangular, FrechetCDF, GeneralizedExtremeValueCDF GeneralizedPareto, GompertzCDF, GumbelCDF, Identity, Inline, Invert IteratedSigmoidCentered, KumaraswamyCDF, LambertWTail, Log, Log1p MaskedAutoregressiveFlow, MatrixInverseTriL, MatvecLU, MoyalCDF, NormalCDF Ordered, Pad, Permute, PowerTransform, RationalQuadraticSpline, RayleighCDF RealNVP, Reciprocal, Reshape, Scale, ScaleMatvecDiag, ScaleMatvecLU ScaleMatvecLinearOperator, ScaleMatvecTriL, ScaleTriL, Shift, ShiftedGompertzCDF Sigmoid, Sinh, SinhArcsinh, SoftClip, Softfloor, SoftmaxCentered, Softplus Softsign, Split, Square, Tanh, TransformDiagonal, Transpose, WeibullCDF
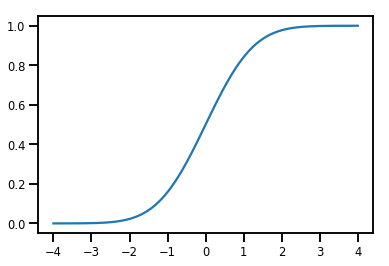
Basit Bijector
normal_cdf = tfp.bijectors.NormalCDF()
xs = np.linspace(-4., 4., 200)
plt.plot(xs, normal_cdf.forward(xs))
plt.show()
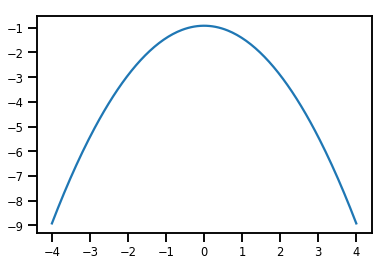
plt.plot(xs, normal_cdf.forward_log_det_jacobian(xs, event_ndims=0))
plt.show()
Bir Bijector bir transforme Distribution
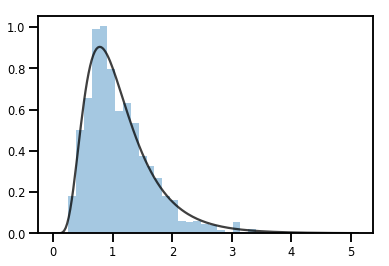
exp_bijector = tfp.bijectors.Exp()
log_normal = exp_bijector(tfd.Normal(0., .5))
samples = log_normal.sample(1000)
xs = np.linspace(1e-10, np.max(samples), 200)
sns.distplot(samples, norm_hist=True, kde=False)
plt.plot(xs, log_normal.prob(xs), c='k', alpha=.75)
plt.show()
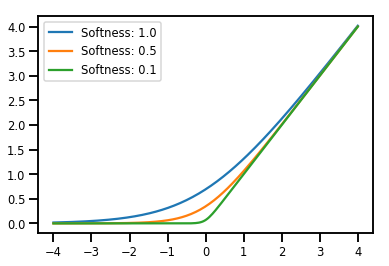
Harmanlama Bijectors
# Create a batch of bijectors of shape [3,]
softplus = tfp.bijectors.Softplus(
hinge_softness=[1., .5, .1])
print("Hinge softness shape:", softplus.hinge_softness.shape)
Hinge softness shape: (3,)
# For broadcasting, we want this to be shape [200, 1]
xs = np.linspace(-4., 4., 200)[..., np.newaxis]
ys = softplus.forward(xs)
print("Forward shape:", ys.shape)
Forward shape: (200, 3)
# Visualization
lines = plt.plot(np.tile(xs, 3), ys)
for line, hs in zip(lines, softplus.hinge_softness):
line.set_label("Softness: %1.1f" % hs)
plt.legend()
plt.show()
Önbelleğe almak
# This bijector represents a matrix outer product on the forward pass,
# and a cholesky decomposition on the inverse pass. The latter costs O(N^3)!
bij = tfb.CholeskyOuterProduct()
size = 2500
# Make a big, lower-triangular matrix
big_lower_triangular = tf.eye(size)
# Squaring it gives us a positive-definite matrix
big_positive_definite = bij.forward(big_lower_triangular)
# Caching for the win!
%timeit bij.inverse(big_positive_definite)
%timeit tf.linalg.cholesky(big_positive_definite)
10000 loops, best of 3: 114 µs per loop 1 loops, best of 3: 208 ms per loop
MCMC
TFP, Hamiltonian Monte Carlo dahil olmak üzere bazı standart Markov zinciri Monte Carlo algoritmaları için destek oluşturmuştur.
Bir veri seti oluşturun
# Generate some data
def f(x, w):
# Pad x with 1's so we can add bias via matmul
x = tf.pad(x, [[1, 0], [0, 0]], constant_values=1)
linop = tf.linalg.LinearOperatorFullMatrix(w[..., np.newaxis])
result = linop.matmul(x, adjoint=True)
return result[..., 0, :]
num_features = 2
num_examples = 50
noise_scale = .5
true_w = np.array([-1., 2., 3.])
xs = np.random.uniform(-1., 1., [num_features, num_examples])
ys = f(xs, true_w) + np.random.normal(0., noise_scale, size=num_examples)
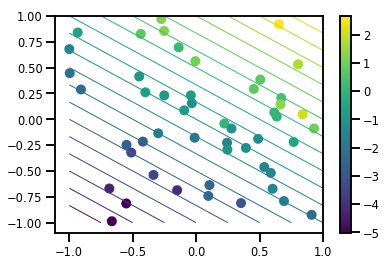
# Visualize the data set
plt.scatter(*xs, c=ys, s=100, linewidths=0)
grid = np.meshgrid(*([np.linspace(-1, 1, 100)] * 2))
xs_grid = np.stack(grid, axis=0)
fs_grid = f(xs_grid.reshape([num_features, -1]), true_w)
fs_grid = np.reshape(fs_grid, [100, 100])
plt.colorbar()
plt.contour(xs_grid[0, ...], xs_grid[1, ...], fs_grid, 20, linewidths=1)
plt.show()
Ortak log-prob fonksiyonumuzu tanımlayın
Normalleştirilmemiş posterior oluşturmak üzere veri üzerinde kapatma sonucudur kısmi uygulaması eklem günlük prob.
# Define the joint_log_prob function, and our unnormalized posterior.
def joint_log_prob(w, x, y):
# Our model in maths is
# w ~ MVN([0, 0, 0], diag([1, 1, 1]))
# y_i ~ Normal(w @ x_i, noise_scale), i=1..N
rv_w = tfd.MultivariateNormalDiag(
loc=np.zeros(num_features + 1),
scale_diag=np.ones(num_features + 1))
rv_y = tfd.Normal(f(x, w), noise_scale)
return (rv_w.log_prob(w) +
tf.reduce_sum(rv_y.log_prob(y), axis=-1))
# Create our unnormalized target density by currying x and y from the joint.
def unnormalized_posterior(w):
return joint_log_prob(w, xs, ys)
HMC TransitionKernel'i oluşturun ve sample_chain'i arayın
# Create an HMC TransitionKernel
hmc_kernel = tfp.mcmc.HamiltonianMonteCarlo(
target_log_prob_fn=unnormalized_posterior,
step_size=np.float64(.1),
num_leapfrog_steps=2)
# We wrap sample_chain in tf.function, telling TF to precompile a reusable
# computation graph, which will dramatically improve performance.
@tf.function
def run_chain(initial_state, num_results=1000, num_burnin_steps=500):
return tfp.mcmc.sample_chain(
num_results=num_results,
num_burnin_steps=num_burnin_steps,
current_state=initial_state,
kernel=hmc_kernel,
trace_fn=lambda current_state, kernel_results: kernel_results)
initial_state = np.zeros(num_features + 1)
samples, kernel_results = run_chain(initial_state)
print("Acceptance rate:", kernel_results.is_accepted.numpy().mean())
Acceptance rate: 0.915
Bu harika değil! .65'e yakın bir kabul oranı istiyoruz.
(bakınız "Optimum Çeşitli Metropolis- Hastings Algorithms Ölçekleme" , Roberts & Rosenthal, 2001)
Uyarlanabilir adım boyutları
Biz bizim HMC TransitionKernel sarar SimpleStepSizeAdaptation Burnin sırasında HMC adım boyutuna adapte bazı (daha doğrusu basit sezgisel) mantığı geçerli olacak olan "meta kernel". Yakmanın %80'ini adım boyutunu uyarlamak için ayırıyoruz ve sonra kalan %20'yi sadece karıştırmaya bırakıyoruz.
# Apply a simple step size adaptation during burnin
@tf.function
def run_chain(initial_state, num_results=1000, num_burnin_steps=500):
adaptive_kernel = tfp.mcmc.SimpleStepSizeAdaptation(
hmc_kernel,
num_adaptation_steps=int(.8 * num_burnin_steps),
target_accept_prob=np.float64(.65))
return tfp.mcmc.sample_chain(
num_results=num_results,
num_burnin_steps=num_burnin_steps,
current_state=initial_state,
kernel=adaptive_kernel,
trace_fn=lambda cs, kr: kr)
samples, kernel_results = run_chain(
initial_state=np.zeros(num_features+1))
print("Acceptance rate:", kernel_results.inner_results.is_accepted.numpy().mean())
Acceptance rate: 0.634
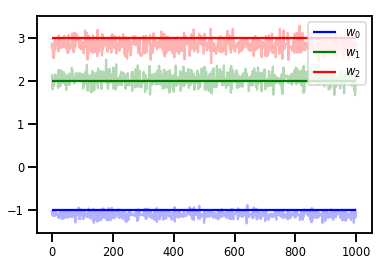
# Trace plots
colors = ['b', 'g', 'r']
for i in range(3):
plt.plot(samples[:, i], c=colors[i], alpha=.3)
plt.hlines(true_w[i], 0, 1000, zorder=4, color=colors[i], label="$w_{}$".format(i))
plt.legend(loc='upper right')
plt.show()
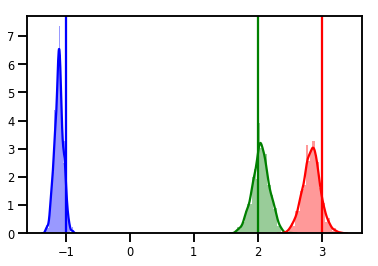
# Histogram of samples
for i in range(3):
sns.distplot(samples[:, i], color=colors[i])
ymax = plt.ylim()[1]
for i in range(3):
plt.vlines(true_w[i], 0, ymax, color=colors[i])
plt.ylim(0, ymax)
plt.show()
teşhis
İz grafikleri güzel, ancak teşhisler daha güzel!
İlk önce birden fazla zincir çalıştırmamız gerekiyor. Bu toplu verme gibi basit gibidir initial_state tensörlerle.
# Instead of a single set of initial w's, we create a batch of 8.
num_chains = 8
initial_state = np.zeros([num_chains, num_features + 1])
chains, kernel_results = run_chain(initial_state)
r_hat = tfp.mcmc.potential_scale_reduction(chains)
print("Acceptance rate:", kernel_results.inner_results.is_accepted.numpy().mean())
print("R-hat diagnostic (per latent variable):", r_hat.numpy())
Acceptance rate: 0.59175 R-hat diagnostic (per latent variable): [0.99998395 0.99932185 0.9997064 ]
Gürültü ölçeğini örnekleme
# Define the joint_log_prob function, and our unnormalized posterior.
def joint_log_prob(w, sigma, x, y):
# Our model in maths is
# w ~ MVN([0, 0, 0], diag([1, 1, 1]))
# y_i ~ Normal(w @ x_i, noise_scale), i=1..N
rv_w = tfd.MultivariateNormalDiag(
loc=np.zeros(num_features + 1),
scale_diag=np.ones(num_features + 1))
rv_sigma = tfd.LogNormal(np.float64(1.), np.float64(5.))
rv_y = tfd.Normal(f(x, w), sigma[..., np.newaxis])
return (rv_w.log_prob(w) +
rv_sigma.log_prob(sigma) +
tf.reduce_sum(rv_y.log_prob(y), axis=-1))
# Create our unnormalized target density by currying x and y from the joint.
def unnormalized_posterior(w, sigma):
return joint_log_prob(w, sigma, xs, ys)
# Create an HMC TransitionKernel
hmc_kernel = tfp.mcmc.HamiltonianMonteCarlo(
target_log_prob_fn=unnormalized_posterior,
step_size=np.float64(.1),
num_leapfrog_steps=4)
# Create a TransformedTransitionKernl
transformed_kernel = tfp.mcmc.TransformedTransitionKernel(
inner_kernel=hmc_kernel,
bijector=[tfb.Identity(), # w
tfb.Invert(tfb.Softplus())]) # sigma
# Apply a simple step size adaptation during burnin
@tf.function
def run_chain(initial_state, num_results=1000, num_burnin_steps=500):
adaptive_kernel = tfp.mcmc.SimpleStepSizeAdaptation(
transformed_kernel,
num_adaptation_steps=int(.8 * num_burnin_steps),
target_accept_prob=np.float64(.75))
return tfp.mcmc.sample_chain(
num_results=num_results,
num_burnin_steps=num_burnin_steps,
current_state=initial_state,
kernel=adaptive_kernel,
seed=(0, 1),
trace_fn=lambda cs, kr: kr)
# Instead of a single set of initial w's, we create a batch of 8.
num_chains = 8
initial_state = [np.zeros([num_chains, num_features + 1]),
.54 * np.ones([num_chains], dtype=np.float64)]
chains, kernel_results = run_chain(initial_state)
r_hat = tfp.mcmc.potential_scale_reduction(chains)
print("Acceptance rate:", kernel_results.inner_results.inner_results.is_accepted.numpy().mean())
print("R-hat diagnostic (per w variable):", r_hat[0].numpy())
print("R-hat diagnostic (sigma):", r_hat[1].numpy())
Acceptance rate: 0.715875 R-hat diagnostic (per w variable): [1.0000073 1.00458208 1.00450512] R-hat diagnostic (sigma): 1.0092056996149859
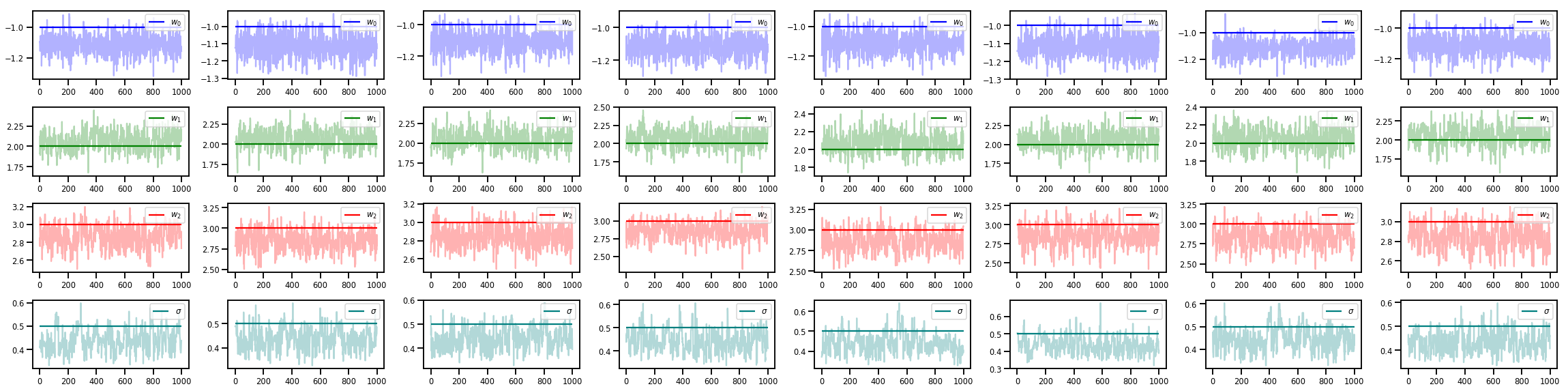
w_chains, sigma_chains = chains
# Trace plots of w (one of 8 chains)
colors = ['b', 'g', 'r', 'teal']
fig, axes = plt.subplots(4, num_chains, figsize=(4 * num_chains, 8))
for j in range(num_chains):
for i in range(3):
ax = axes[i][j]
ax.plot(w_chains[:, j, i], c=colors[i], alpha=.3)
ax.hlines(true_w[i], 0, 1000, zorder=4, color=colors[i], label="$w_{}$".format(i))
ax.legend(loc='upper right')
ax = axes[3][j]
ax.plot(sigma_chains[:, j], alpha=.3, c=colors[3])
ax.hlines(noise_scale, 0, 1000, zorder=4, color=colors[3], label=r"$\sigma$".format(i))
ax.legend(loc='upper right')
fig.tight_layout()
plt.show()
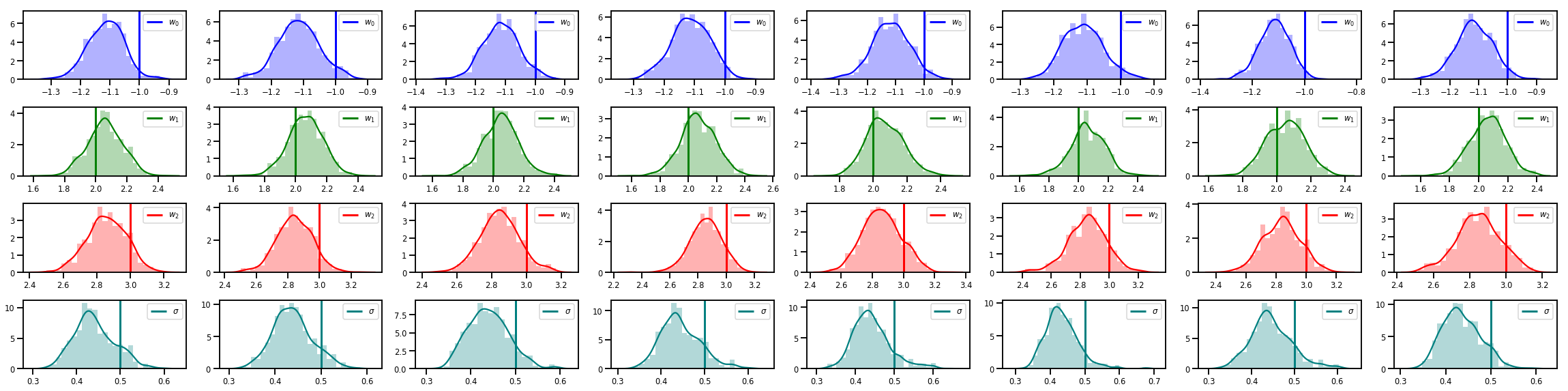
# Histogram of samples of w
fig, axes = plt.subplots(4, num_chains, figsize=(4 * num_chains, 8))
for j in range(num_chains):
for i in range(3):
ax = axes[i][j]
sns.distplot(w_chains[:, j, i], color=colors[i], norm_hist=True, ax=ax, hist_kws={'alpha': .3})
for i in range(3):
ax = axes[i][j]
ymax = ax.get_ylim()[1]
ax.vlines(true_w[i], 0, ymax, color=colors[i], label="$w_{}$".format(i), linewidth=3)
ax.set_ylim(0, ymax)
ax.legend(loc='upper right')
ax = axes[3][j]
sns.distplot(sigma_chains[:, j], color=colors[3], norm_hist=True, ax=ax, hist_kws={'alpha': .3})
ymax = ax.get_ylim()[1]
ax.vlines(noise_scale, 0, ymax, color=colors[3], label=r"$\sigma$".format(i), linewidth=3)
ax.set_ylim(0, ymax)
ax.legend(loc='upper right')
fig.tight_layout()
plt.show()
Çok daha fazlası var!
Bu harika blog gönderilerine ve örneklere göz atın:
- Yapısal Zaman Serisi desteği günlüğü CoLab
- Olasılık Keras Katmanlar (giriş: tensör, çıktı: Dağıtım!) Blog CoLab
- Gauss işlem Regresyon CoLab ve latent değişken Modelleme CoLab
Bizim GitHub üzerinde fazla örnek ve dizüstü bilgisayarlar burada !
