Zamiarem tego notebooka jest pomóc TFP 0.13.0 „ożyć” za pomocą kilku małych fragmentów – małych demonstracji rzeczy, które można osiągnąć za pomocą TFP.
 Zobacz na TensorFlow.org Zobacz na TensorFlow.org |  Uruchom w Google Colab Uruchom w Google Colab |  Wyświetl źródło na GitHub Wyświetl źródło na GitHub |  Pobierz notatnik Pobierz notatnik |
Instalacje i importy
!pip3 install -qU tensorflow==2.5.0 tensorflow_probability==0.13.0 tensorflow-datasets inference_gym
import tensorflow as tf
import tensorflow_probability as tfp
assert '0.13' in tfp.__version__, tfp.__version__
assert '2.5' in tf.__version__, tf.__version__
physical_devices = tf.config.list_physical_devices('CPU')
tf.config.set_logical_device_configuration(
physical_devices[0],
[tf.config.LogicalDeviceConfiguration(),
tf.config.LogicalDeviceConfiguration()])
tfd = tfp.distributions
tfb = tfp.bijectors
tfpk = tfp.math.psd_kernels
import matplotlib.pyplot as plt
import numpy as np
import scipy.interpolate
import IPython
import seaborn as sns
import logging
[K |████████████████████████████████| 5.4MB 8.8MB/s [K |████████████████████████████████| 3.9MB 37.1MB/s [K |████████████████████████████████| 296kB 31.6MB/s [?25h
Dystrybucje [matematyka podstawowa]
BetaQuotient
Stosunek dwóch niezależnych zmiennych losowych o rozkładzie beta
plt.hist(tfd.BetaQuotient(concentration1_numerator=5.,
concentration0_numerator=2.,
concentration1_denominator=3.,
concentration0_denominator=8.).sample(1_000, seed=(1, 23)),
bins='auto');
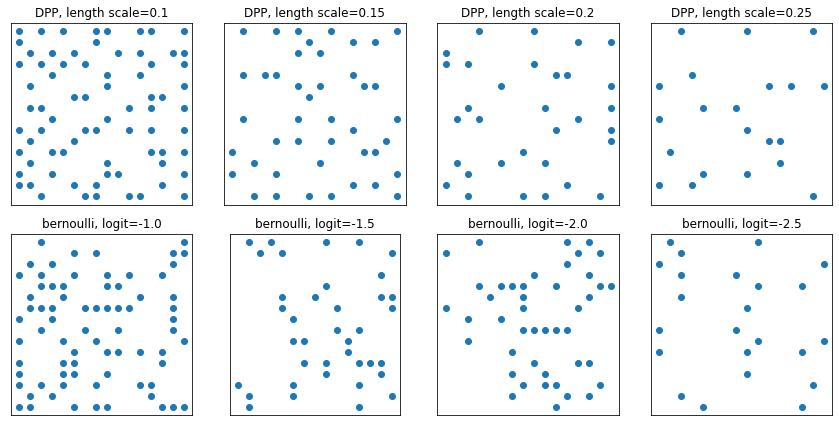
DeterminantalPointProcess
Rozkład na podzbiory (reprezentowane jako jeden gorący) danego zbioru. Próbki mają właściwość odpychania (prawdopodobieństwa są proporcjonalne do objętości rozpiętej przez wektory odpowiadające wybranemu podzbiorowi punktów), która ma tendencję do próbkowania różnych podzbiorów. [Porównaj z próbkami iid Bernoulliego.]
grid_size = 16
# Generate grid_size**2 pts on the unit square.
grid = np.arange(0, 1, 1./grid_size).astype(np.float32)
import itertools
points = np.array(list(itertools.product(grid, grid)))
# Create the kernel L that parameterizes the DPP.
kernel_amplitude = 2.
kernel_lengthscale = [.1, .15, .2, .25] # Increasing length scale indicates more points are "nearby", tending toward smaller subsets.
kernel = tfpk.ExponentiatedQuadratic(kernel_amplitude, kernel_lengthscale)
kernel_matrix = kernel.matrix(points, points)
eigenvalues, eigenvectors = tf.linalg.eigh(kernel_matrix)
dpp = tfd.DeterminantalPointProcess(eigenvalues, eigenvectors)
print(dpp)
# The inner-most dimension of the result of `dpp.sample` is a multi-hot
# encoding of a subset of {1, ..., ground_set_size}.
# We will compare against a bernoulli distribution.
samps_dpp = dpp.sample(seed=(1, 2)) # 4 x grid_size**2
logits = tf.broadcast_to([[-1.], [-1.5], [-2], [-2.5]], [4, grid_size**2])
samps_bern = tfd.Bernoulli(logits=logits).sample(seed=(2, 3))
plt.figure(figsize=(12, 6))
for i, (samp, samp_bern) in enumerate(zip(samps_dpp, samps_bern)):
plt.subplot(241 + i)
plt.scatter(*points[np.where(samp)].T)
plt.title(f'DPP, length scale={kernel_lengthscale[i]}')
plt.xticks([])
plt.yticks([])
plt.gca().set_aspect(1.)
plt.subplot(241 + i + 4)
plt.scatter(*points[np.where(samp_bern)].T)
plt.title(f'bernoulli, logit={logits[i,0]}')
plt.xticks([])
plt.yticks([])
plt.gca().set_aspect(1.)
plt.tight_layout()
plt.show()
tfp.distributions.DeterminantalPointProcess("DeterminantalPointProcess", batch_shape=[4], event_shape=[256], dtype=int32)
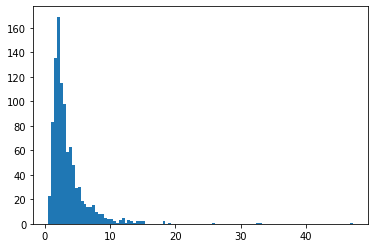
SigmoidBeta
Logarytmiczne szanse dwóch rozkładów gamma. Więcej numerycznie stabilny przestrzeń próbka niż Beta .
plt.hist(tfd.SigmoidBeta(concentration1=.01, concentration0=2.).sample(10_000, seed=(1, 23)),
bins='auto', density=True);
plt.show()
print('Old way, fractions non-finite:')
print(np.sum(~tf.math.is_finite(
tfb.Invert(tfb.Sigmoid())(tfd.Beta(concentration1=.01, concentration0=2.)).sample(10_000, seed=(1, 23)))) / 10_000)
print(np.sum(~tf.math.is_finite(
tfb.Invert(tfb.Sigmoid())(tfd.Beta(concentration1=2., concentration0=.01)).sample(10_000, seed=(2, 34)))) / 10_000)
Old way, fractions non-finite: 0.4215 0.8624
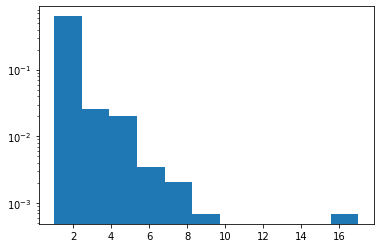
Zipf
Dodano obsługę JAX.
plt.hist(tfd.Zipf(3.).sample(1_000, seed=(12, 34)).numpy(), bins='auto', density=True, log=True);
NormalInverseGaussian
Elastyczna rodzina parametryczna obsługująca ciężkie ogony, skośne i waniliowe Normal.
MatrixNormalLinearOperator
Macierz Rozkład normalny.
# Initialize a single 2 x 3 Matrix Normal.
mu = [[1., 2, 3], [3., 4, 5]]
col_cov = [[ 0.36, 0.12, 0.06],
[ 0.12, 0.29, -0.13],
[ 0.06, -0.13, 0.26]]
scale_column = tf.linalg.LinearOperatorLowerTriangular(tf.linalg.cholesky(col_cov))
scale_row = tf.linalg.LinearOperatorDiag([0.9, 0.8])
mvn = tfd.MatrixNormalLinearOperator(loc=mu, scale_row=scale_row, scale_column=scale_column)
mvn.sample()
WARNING:tensorflow:From /usr/local/lib/python3.7/dist-packages/tensorflow/python/ops/linalg/linear_operator_kronecker.py:224: LinearOperator.graph_parents (from tensorflow.python.ops.linalg.linear_operator) is deprecated and will be removed in a future version.
Instructions for updating:
Do not call `graph_parents`.
<tf.Tensor: shape=(2, 3), dtype=float32, numpy=
array([[1.2495145, 1.549366 , 3.2748342],
[3.7330258, 4.3413105, 4.83423 ]], dtype=float32)>
MatrixStudentTLinearOperator
Rozkład macierzy T.
mu = [[1., 2, 3], [3., 4, 5]]
col_cov = [[ 0.36, 0.12, 0.06],
[ 0.12, 0.29, -0.13],
[ 0.06, -0.13, 0.26]]
scale_column = tf.linalg.LinearOperatorLowerTriangular(tf.linalg.cholesky(col_cov))
scale_row = tf.linalg.LinearOperatorDiag([0.9, 0.8])
mvn = tfd.MatrixTLinearOperator(
df=2.,
loc=mu,
scale_row=scale_row,
scale_column=scale_column)
mvn.sample()
<tf.Tensor: shape=(2, 3), dtype=float32, numpy=
array([[1.6549466, 2.6708362, 2.8629923],
[2.1222284, 3.6904747, 5.08014 ]], dtype=float32)>
Dystrybucje [oprogramowanie / wrappery]
Sharded
Odłamuje niezależne fragmenty zdarzeń dystrybucji na wiele procesorów. Kruszywa log_prob różnych urządzeniach, uchwyty gradienty w porozumieniu z tfp.experimental.distribute.JointDistribution* . Znacznie więcej w Ukazuje Inference notebooka.
strategy = tf.distribute.MirroredStrategy()
@tf.function
def sample_and_lp(seed):
d = tfp.experimental.distribute.Sharded(tfd.Normal(0, 1))
s = d.sample(seed=seed)
return s, d.log_prob(s)
strategy.run(sample_and_lp, args=(tf.constant([12,34]),))
WARNING:tensorflow:There are non-GPU devices in `tf.distribute.Strategy`, not using nccl allreduce.
WARNING:tensorflow:Collective ops is not configured at program startup. Some performance features may not be enabled.
INFO:tensorflow:Using MirroredStrategy with devices ('/job:localhost/replica:0/task:0/device:CPU:0', '/job:localhost/replica:0/task:0/device:CPU:1')
INFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0', '/job:localhost/replica:0/task:0/device:CPU:1').
(PerReplica:{
0: <tf.Tensor: shape=(), dtype=float32, numpy=0.0051413667>,
1: <tf.Tensor: shape=(), dtype=float32, numpy=-0.3393052>
}, PerReplica:{
0: <tf.Tensor: shape=(), dtype=float32, numpy=-1.8954543>,
1: <tf.Tensor: shape=(), dtype=float32, numpy=-1.8954543>
})
BatchBroadcast
Niejawnie nadawanie wymiarów wsad dystrybucji bazowego z lub do danego kształtu wsadowym.
underlying = tfd.MultivariateNormalDiag(tf.zeros([7, 1, 5]), tf.ones([5]))
print('underlying:', underlying)
d = tfd.BatchBroadcast(underlying, [8, 1, 6])
print('broadcast [7, 1] *with* [8, 1, 6]:', d)
try:
tfd.BatchBroadcast(underlying, to_shape=[8, 1, 6])
except ValueError as e:
print('broadcast [7, 1] *to* [8, 1, 6] is invalid:', e)
d = tfd.BatchBroadcast(underlying, to_shape=[8, 7, 6])
print('broadcast [7, 1] *to* [8, 7, 6]:', d)
underlying: tfp.distributions.MultivariateNormalDiag("MultivariateNormalDiag", batch_shape=[7, 1], event_shape=[5], dtype=float32)
broadcast [7, 1] *with* [8, 1, 6]: tfp.distributions.BatchBroadcast("BatchBroadcastMultivariateNormalDiag", batch_shape=[8, 7, 6], event_shape=[5], dtype=float32)
broadcast [7, 1] *to* [8, 1, 6] is invalid: Argument `to_shape` ([8 1 6]) is incompatible with underlying distribution batch shape ((7, 1)).
broadcast [7, 1] *to* [8, 7, 6]: tfp.distributions.BatchBroadcast("BatchBroadcastMultivariateNormalDiag", batch_shape=[8, 7, 6], event_shape=[5], dtype=float32)
Masked
Dla pojedynczego programu / wielu danych jednostkowych lub rozrzedzony-as-zamaskowanych gęsty użytkowych przypadkach, dystrybucji, maski na zewnątrz log_prob nieprawidłowych wypłat bazowych.
d = tfd.Masked(tfd.Normal(tf.zeros([7]), 1),
validity_mask=tf.sequence_mask([3, 4], 7))
print(d.log_prob(d.sample(seed=(1, 1))))
d = tfd.Masked(tfd.Normal(0, 1),
validity_mask=[False, True, False],
safe_sample_fn=tfd.Distribution.mode)
print(d.log_prob(d.sample(seed=(2, 2))))
tf.Tensor( [[-2.3054113 -1.8524303 -1.2220721 0. 0. 0. 0. ] [-1.118623 -1.1370811 -1.1574132 -5.884986 0. 0. 0. ]], shape=(2, 7), dtype=float32) tf.Tensor([ 0. -0.93683904 0. ], shape=(3,), dtype=float32)
Bijektory
- Bijektory
- Dodaj bijectors naśladować
tf.nest.flatten(tfb.tree_flatten) itf.nest.pack_sequence_as(tfb.pack_sequence_as). - Dodaje
tfp.experimental.bijectors.Sharded - Usunąć przestarzałe
tfb.ScaleTrilL. Zastosowanietfb.FillScaleTriLzamiast. - Dodaje
cls.parameter_properties()adnotacje dotyczące Bijectors. - Rozszerzyć zakres
tfb.Powerdla wszystkich liczb rzeczywistych dla nieparzystych potęg całkowitych. - Wywnioskuj log-deg-jacobian bijektorów skalarnych przy użyciu autodiff, jeśli nie określono inaczej.
- Dodaj bijectors naśladować
Restrukturyzacyjne bijektory
ex = (tf.constant(1.), dict(b=tf.constant(2.), c=tf.constant(3.)))
b = tfb.tree_flatten(ex)
print(b.forward(ex))
print(b.inverse(list(tf.constant([1., 2, 3]))))
b = tfb.pack_sequence_as(ex)
print(b.forward(list(tf.constant([1., 2, 3]))))
print(b.inverse(ex))
[<tf.Tensor: shape=(), dtype=float32, numpy=1.0>, <tf.Tensor: shape=(), dtype=float32, numpy=2.0>, <tf.Tensor: shape=(), dtype=float32, numpy=3.0>]
(<tf.Tensor: shape=(), dtype=float32, numpy=1.0>, {'b': <tf.Tensor: shape=(), dtype=float32, numpy=2.0>, 'c': <tf.Tensor: shape=(), dtype=float32, numpy=3.0>})
(<tf.Tensor: shape=(), dtype=float32, numpy=1.0>, {'b': <tf.Tensor: shape=(), dtype=float32, numpy=2.0>, 'c': <tf.Tensor: shape=(), dtype=float32, numpy=3.0>})
[<tf.Tensor: shape=(), dtype=float32, numpy=1.0>, <tf.Tensor: shape=(), dtype=float32, numpy=2.0>, <tf.Tensor: shape=(), dtype=float32, numpy=3.0>]
Sharded
Redukcja SPMD w wyznaczniku logarytmicznym. Zobacz Sharded w rozkładzie poniżej.
strategy = tf.distribute.MirroredStrategy()
def sample_lp_logdet(seed):
d = tfd.TransformedDistribution(tfp.experimental.distribute.Sharded(tfd.Normal(0, 1), shard_axis_name='i'),
tfp.experimental.bijectors.Sharded(tfb.Sigmoid(), shard_axis_name='i'))
s = d.sample(seed=seed)
return s, d.log_prob(s), d.bijector.inverse_log_det_jacobian(s)
strategy.run(sample_lp_logdet, (tf.constant([1, 2]),))
WARNING:tensorflow:There are non-GPU devices in `tf.distribute.Strategy`, not using nccl allreduce.
WARNING:tensorflow:Collective ops is not configured at program startup. Some performance features may not be enabled.
INFO:tensorflow:Using MirroredStrategy with devices ('/job:localhost/replica:0/task:0/device:CPU:0', '/job:localhost/replica:0/task:0/device:CPU:1')
WARNING:tensorflow:Using MirroredStrategy eagerly has significant overhead currently. We will be working on improving this in the future, but for now please wrap `call_for_each_replica` or `experimental_run` or `run` inside a tf.function to get the best performance.
INFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0', '/job:localhost/replica:0/task:0/device:CPU:1').
INFO:tensorflow:Reduce to /job:localhost/replica:0/task:0/device:CPU:0 then broadcast to ('/job:localhost/replica:0/task:0/device:CPU:0', '/job:localhost/replica:0/task:0/device:CPU:1').
(PerReplica:{
0: <tf.Tensor: shape=(), dtype=float32, numpy=0.87746525>,
1: <tf.Tensor: shape=(), dtype=float32, numpy=0.24580425>
}, PerReplica:{
0: <tf.Tensor: shape=(), dtype=float32, numpy=-0.48870325>,
1: <tf.Tensor: shape=(), dtype=float32, numpy=-0.48870325>
}, PerReplica:{
0: <tf.Tensor: shape=(), dtype=float32, numpy=3.9154015>,
1: <tf.Tensor: shape=(), dtype=float32, numpy=3.9154015>
})
VI
- Dodaje
build_split_flow_surrogate_posteriordotfp.experimental.vizbudować strukturę VI zastępczych tyłki z przepływów normalizujące. - Dodaje
build_affine_surrogate_posteriordotfp.experimental.vina budowę ADVI posteriors zastępczych z kształtu zdarzeń. - Dodaje
build_affine_surrogate_posterior_from_base_distributiondotfp.experimental.viaby umożliwić budowę posteriors ADVI zastępczych ze strukturami korelacji indukowanych przez transformacje afiniczne.
VI/MAP/MLE
- Dodano metoda wygoda
tfp.experimental.util.make_trainable(cls), aby utworzyć wyszkolić wystąpień dystrybucji i bijectors.
d = tfp.experimental.util.make_trainable(tfd.Gamma)
print(d.trainable_variables)
print(d)
(<tf.Variable 'Gamma_trainable_variables/concentration:0' shape=() dtype=float32, numpy=1.0296053>, <tf.Variable 'Gamma_trainable_variables/log_rate:0' shape=() dtype=float32, numpy=-0.3465951>)
tfp.distributions.Gamma("Gamma", batch_shape=[], event_shape=[], dtype=float32)
MCMC
- Diagnostyka MCMC obsługuje dowolne struktury stanów, a nie tylko listy.
-
remc_thermodynamic_integralsdodano dotfp.experimental.mcmc - Dodaje
tfp.experimental.mcmc.windowed_adaptive_hmc - Dodaje eksperymentalny interfejs API do inicjowania łańcucha Markowa z niemal zerowej jednolitej dystrybucji w nieograniczonej przestrzeni.
tfp.experimental.mcmc.init_near_unconstrained_zero - Dodaje eksperymentalne narzędzie do ponawiania próby inicjalizacji łańcucha Markowa do momentu znalezienia akceptowalnego punktu.
tfp.experimental.mcmc.retry_init - Tasowanie eksperymentalnego interfejsu API MCMC do przesyłania strumieniowego w celu umieszczenia go w tfp.mcmc z minimalnymi zakłóceniami.
- Dodaje
ThinningKerneldoexperimental.mcmc. - Dodaje
experimental.mcmc.run_kernelkierowca jako kandydata strumieniowe oparte na zastępstwomcmc.sample_chain
init_near_unconstrained_zero , retry_init
@tfd.JointDistributionCoroutine
def model():
Root = tfd.JointDistributionCoroutine.Root
c0 = yield Root(tfd.Gamma(2, 2, name='c0'))
c1 = yield Root(tfd.Gamma(2, 2, name='c1'))
counts = yield tfd.Sample(tfd.BetaBinomial(23, c1, c0), 10, name='counts')
jd = model.experimental_pin(counts=model.sample(seed=[20, 30]).counts)
init_dist = tfp.experimental.mcmc.init_near_unconstrained_zero(jd)
print(init_dist)
tfp.experimental.mcmc.retry_init(init_dist.sample, jd.unnormalized_log_prob)
tfp.distributions.TransformedDistribution("default_joint_bijectorrestructureJointDistributionSequential", batch_shape=StructTuple(
c0=[],
c1=[]
), event_shape=StructTuple(
c0=[],
c1=[]
), dtype=StructTuple(
c0=float32,
c1=float32
))
StructTuple(
c0=<tf.Tensor: shape=(), dtype=float32, numpy=1.7879653>,
c1=<tf.Tensor: shape=(), dtype=float32, numpy=0.34548905>
)
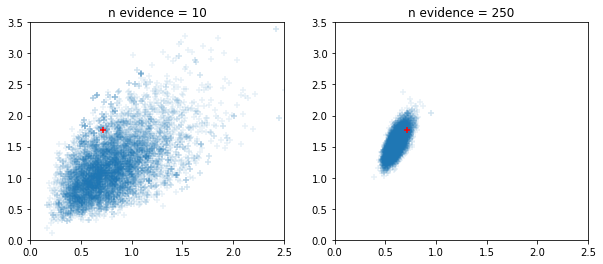
Okienkowe adaptacyjne próbniki HMC i NUTS
fig, ax = plt.subplots(1, 2, figsize=(10, 4))
for i, n_evidence in enumerate((10, 250)):
ax[i].set_title(f'n evidence = {n_evidence}')
ax[i].set_xlim(0, 2.5); ax[i].set_ylim(0, 3.5)
@tfd.JointDistributionCoroutine
def model():
Root = tfd.JointDistributionCoroutine.Root
c0 = yield Root(tfd.Gamma(2, 2, name='c0'))
c1 = yield Root(tfd.Gamma(2, 2, name='c1'))
counts = yield tfd.Sample(tfd.BetaBinomial(23, c1, c0), n_evidence, name='counts')
s = model.sample(seed=[20, 30])
print(s)
jd = model.experimental_pin(counts=s.counts)
states, trace = tf.function(tfp.experimental.mcmc.windowed_adaptive_hmc)(
100, jd, num_leapfrog_steps=5, seed=[100, 200])
ax[i].scatter(states.c0.numpy().reshape(-1), states.c1.numpy().reshape(-1),
marker='+', alpha=.1)
ax[i].scatter(s.c0, s.c1, marker='+', color='r')
StructTuple(
c0=<tf.Tensor: shape=(), dtype=float32, numpy=0.7161876>,
c1=<tf.Tensor: shape=(), dtype=float32, numpy=1.7696666>,
counts=<tf.Tensor: shape=(10,), dtype=float32, numpy=array([ 6., 10., 23., 7., 2., 20., 14., 16., 22., 17.], dtype=float32)>
)
WARNING:tensorflow:6 out of the last 6 calls to <function windowed_adaptive_hmc at 0x7fda42bed8c0> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
StructTuple(
c0=<tf.Tensor: shape=(), dtype=float32, numpy=0.7161876>,
c1=<tf.Tensor: shape=(), dtype=float32, numpy=1.7696666>,
counts=<tf.Tensor: shape=(250,), dtype=float32, numpy=
array([ 6., 10., 23., 7., 2., 20., 14., 16., 22., 17., 22., 21., 6.,
21., 12., 22., 23., 16., 18., 21., 16., 17., 17., 16., 21., 14.,
23., 15., 10., 19., 8., 23., 23., 14., 1., 23., 16., 22., 20.,
20., 22., 15., 16., 20., 20., 21., 23., 22., 21., 15., 18., 23.,
12., 16., 19., 23., 18., 5., 22., 22., 22., 18., 12., 17., 17.,
16., 8., 22., 20., 23., 3., 12., 14., 18., 7., 19., 19., 9.,
10., 23., 14., 22., 22., 21., 13., 23., 14., 23., 10., 17., 23.,
17., 20., 16., 20., 19., 14., 0., 17., 22., 12., 2., 17., 15.,
14., 23., 19., 15., 23., 2., 21., 23., 21., 7., 21., 12., 23.,
17., 17., 4., 22., 16., 14., 19., 19., 20., 6., 16., 14., 18.,
21., 12., 21., 21., 22., 2., 19., 11., 6., 19., 1., 23., 23.,
14., 6., 23., 18., 8., 20., 23., 13., 20., 18., 23., 17., 22.,
23., 20., 18., 22., 16., 23., 9., 22., 21., 16., 20., 21., 16.,
23., 7., 13., 23., 19., 3., 13., 23., 23., 13., 19., 23., 20.,
18., 8., 19., 14., 12., 6., 8., 23., 3., 13., 21., 23., 22.,
23., 19., 22., 21., 15., 22., 21., 21., 23., 9., 19., 20., 23.,
11., 23., 14., 23., 14., 21., 21., 10., 23., 9., 13., 1., 8.,
8., 20., 21., 21., 21., 14., 16., 16., 9., 23., 22., 11., 23.,
12., 18., 1., 23., 9., 3., 21., 21., 23., 22., 18., 23., 16.,
3., 11., 16.], dtype=float32)>
)
Matematyka, statystyki
Matematyka/linalg
- Dodaj
tfp.math.trapzo trapezoidalnym integracji. - Dodaj
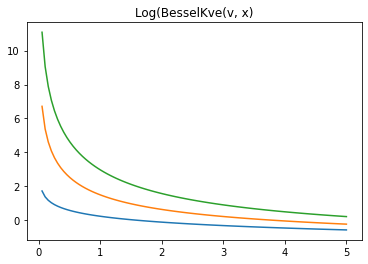
tfp.math.log_bessel_kve. - Dodaj
no_pivot_ldldoexperimental.linalg. - Dodaj
marginal_fnargumentGaussianProcess(patrzno_pivot_ldl). - Dodano
tfp.math.atan_difference(x, y) - Dodaj
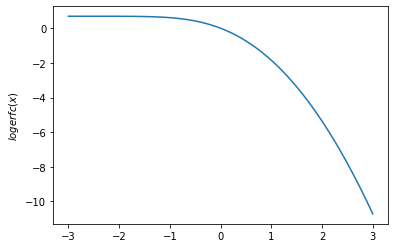
tfp.math.erfcx,tfp.math.logerfcitfp.math.logerfcx - Dodaj
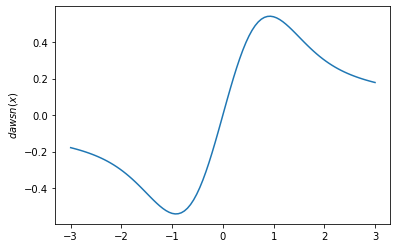
tfp.math.dawsnza integralną Dawsona. - Dodaj
tfp.math.igammaincinv,tfp.math.igammacinv. - Dodaj
tfp.math.sqrt1pm1. - Dodaj
LogitNormal.stddev_approxiLogitNormal.variance_approx - Dodaj
tfp.math.owens_tdo funkcji Owena T. - Dodaj
bracket_rootmetodę automatycznie zainicjować granicach do poszukiwania korzeni. - Dodaj metodę Chandrupatli do znajdowania pierwiastków funkcji skalarnych.
- Dodaj
Statystyki
-
tfp.stats.windowed_meansprawnie wylicza okienkowym środków. -
tfp.stats.windowed_variancesprawnie i dokładnie Oblicza okienkiem wariancje. -
tfp.stats.cumulative_variancewydajnie i precyzyjnie oblicza skumulowane wariancji. -
RunningCovariancei znajomi mogą teraz zostać zainicjowana z przykładu Tensor, nie tylko z wyraźnego kształtu i dtype. - Cleaner API dla
RunningCentralMoments,RunningMean,RunningPotentialScaleReduction.
-
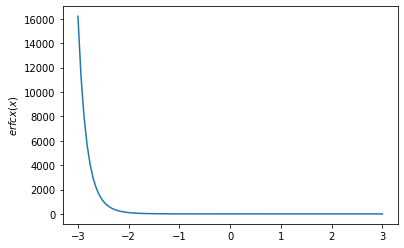
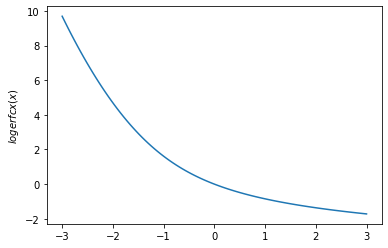
Funkcje Owena T, Erfcx, Logerfc, Logerfcx, Dawson
# Owen's T gives the probability that X > h, 0 < Y < a * X. Let's check that
# with random sampling.
h = np.array([1., 2.]).astype(np.float32)
a = np.array([10., 11.5]).astype(np.float32)
probs = tfp.math.owens_t(h, a)
x = tfd.Normal(0., 1.).sample(int(1e5), seed=(6, 245)).numpy()
y = tfd.Normal(0., 1.).sample(int(1e5), seed=(7, 245)).numpy()
true_values = (
(x[..., np.newaxis] > h) &
(0. < y[..., np.newaxis]) &
(y[..., np.newaxis] < a * x[..., np.newaxis]))
print('Calculated values: {}'.format(
np.count_nonzero(true_values, axis=0) / 1e5))
print('Expected values: {}'.format(probs))
Calculated values: [0.07896 0.01134] Expected values: [0.07932763 0.01137507]
x = np.linspace(-3., 3., 100)
plt.plot(x, tfp.math.erfcx(x))
plt.ylabel('$erfcx(x)$')
plt.show()
plt.plot(x, tfp.math.logerfcx(x))
plt.ylabel('$logerfcx(x)$')
plt.show()
plt.plot(x, tfp.math.logerfc(x))
plt.ylabel('$logerfc(x)$')
plt.show()
plt.plot(x, tfp.math.dawsn(x))
plt.ylabel('$dawsn(x)$')
plt.show()
igammainv / igammacinv
# Igammainv and Igammacinv are inverses to Igamma and Igammac
x = np.linspace(1., 10., 10)
y = tf.math.igamma(0.3, x)
x_prime = tfp.math.igammainv(0.3, y)
print('x: {}'.format(x))
print('igammainv(igamma(a, x)):\n {}'.format(x_prime))
y = tf.math.igammac(0.3, x)
x_prime = tfp.math.igammacinv(0.3, y)
print('\n')
print('x: {}'.format(x))
print('igammacinv(igammac(a, x)):\n {}'.format(x_prime))
x: [ 1. 2. 3. 4. 5. 6. 7. 8. 9. 10.] igammainv(igamma(a, x)): [1. 1.9999992 3.000003 4.0000024 5.0000257 5.999887 7.0002484 7.999243 8.99872 9.994673 ] x: [ 1. 2. 3. 4. 5. 6. 7. 8. 9. 10.] igammacinv(igammac(a, x)): [1. 2. 3. 4. 5. 6. 7. 8.000001 9. 9.999999]
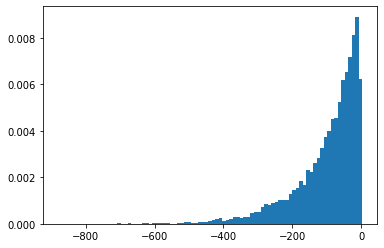
log-kve
x = np.linspace(0., 5., 100)
for v in [0.5, 2., 3]:
plt.plot(x, tfp.math.log_bessel_kve(v, x).numpy())
plt.title('Log(BesselKve(v, x)')
Text(0.5, 1.0, 'Log(BesselKve(v, x)')
Inny
STS
- Przyspieszenie STS prognozowania i rozkładu przy użyciu wewnętrznego
tf.functionopakowania. - Dodaj opcję, aby przyspieszyć filtrowania w
LinearGaussianSSMgdy wymagane są wyniki tylko Ostatnim krokiem jest. - Wariacyjna Wnioskowanie ze wspólnych rozkładów: przykład notebook z modelem Radon .
- Dodaj eksperymentalne wsparcie dla przekształcenia dowolnej dystrybucji w bijektora wstępnego kondycjonowania.
- Przyspieszenie STS prognozowania i rozkładu przy użyciu wewnętrznego
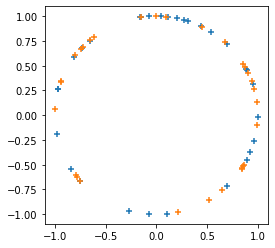
Dodaje
tfp.random.sanitize_seed.Dodaje
tfp.random.spherical_uniform.
plt.figure(figsize=(4, 4))
seed = tfp.random.sanitize_seed(123)
seed1, seed2 = tfp.random.split_seed(seed)
samps = tfp.random.spherical_uniform([30], dimension=2, seed=seed1)
plt.scatter(*samps.numpy().T, marker='+')
samps = tfp.random.spherical_uniform([30], dimension=2, seed=seed2)
plt.scatter(*samps.numpy().T, marker='+');