 View on TensorFlow.org
View on TensorFlow.org
|
 Run in Google Colab
Run in Google Colab
|
 View source on GitHub
View source on GitHub
|
 Download notebook Download notebook
|
This tutorial introduces autoencoders with three examples: the basics, image denoising, and anomaly detection.
An autoencoder is a special type of neural network that is trained to copy its input to its output. For example, given an image of a handwritten digit, an autoencoder first encodes the image into a lower dimensional latent representation, then decodes the latent representation back to an image. An autoencoder learns to compress the data while minimizing the reconstruction error.
To learn more about autoencoders, please consider reading chapter 14 from Deep Learning by Ian Goodfellow, Yoshua Bengio, and Aaron Courville.
Import TensorFlow and other libraries
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import tensorflow as tf
from sklearn.metrics import accuracy_score, precision_score, recall_score
from sklearn.model_selection import train_test_split
from tensorflow.keras import layers, losses
from tensorflow.keras.datasets import fashion_mnist
from tensorflow.keras.models import Model
2024-08-16 05:08:24.454969: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:485] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered 2024-08-16 05:08:24.475836: E external/local_xla/xla/stream_executor/cuda/cuda_dnn.cc:8454] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered 2024-08-16 05:08:24.482184: E external/local_xla/xla/stream_executor/cuda/cuda_blas.cc:1452] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
Load the dataset
To start, you will train the basic autoencoder using the Fashion MNIST dataset. Each image in this dataset is 28x28 pixels.
(x_train, _), (x_test, _) = fashion_mnist.load_data()
x_train = x_train.astype('float32') / 255.
x_test = x_test.astype('float32') / 255.
print (x_train.shape)
print (x_test.shape)
(60000, 28, 28) (10000, 28, 28)
First example: Basic autoencoder
Define an autoencoder with two Dense layers: an encoder, which compresses the images into a 64 dimensional latent vector, and a decoder, that reconstructs the original image from the latent space.
To define your model, use the Keras Model Subclassing API.
class Autoencoder(Model):
def __init__(self, latent_dim, shape):
super(Autoencoder, self).__init__()
self.latent_dim = latent_dim
self.shape = shape
self.encoder = tf.keras.Sequential([
layers.Flatten(),
layers.Dense(latent_dim, activation='relu'),
])
self.decoder = tf.keras.Sequential([
layers.Dense(tf.math.reduce_prod(shape).numpy(), activation='sigmoid'),
layers.Reshape(shape)
])
def call(self, x):
encoded = self.encoder(x)
decoded = self.decoder(encoded)
return decoded
shape = x_test.shape[1:]
latent_dim = 64
autoencoder = Autoencoder(latent_dim, shape)
WARNING: All log messages before absl::InitializeLog() is called are written to STDERR I0000 00:00:1723784907.495092 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784907.498985 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784907.502627 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784907.506226 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784907.518853 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784907.522303 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784907.525756 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784907.529169 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784907.532506 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784907.536023 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784907.539501 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784907.542895 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.772373 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.774589 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.776554 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.778641 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.780679 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.782717 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.784577 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.786661 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.788966 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.791004 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.792858 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.795056 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.833540 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.835672 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.837578 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.839614 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.841487 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.843549 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.845399 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.847401 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.849242 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.851783 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.854229 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723784908.856641 160375 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
autoencoder.compile(optimizer='adam', loss=losses.MeanSquaredError())
Train the model using x_train as both the input and the target. The encoder will learn to compress the dataset from 784 dimensions to the latent space, and the decoder will learn to reconstruct the original images.
.
autoencoder.fit(x_train, x_train,
epochs=10,
shuffle=True,
validation_data=(x_test, x_test))
Epoch 1/10 WARNING: All log messages before absl::InitializeLog() is called are written to STDERR I0000 00:00:1723784911.543932 160540 service.cc:146] XLA service 0x7f8910006f20 initialized for platform CUDA (this does not guarantee that XLA will be used). Devices: I0000 00:00:1723784911.543965 160540 service.cc:154] StreamExecutor device (0): Tesla T4, Compute Capability 7.5 I0000 00:00:1723784911.543969 160540 service.cc:154] StreamExecutor device (1): Tesla T4, Compute Capability 7.5 I0000 00:00:1723784911.543973 160540 service.cc:154] StreamExecutor device (2): Tesla T4, Compute Capability 7.5 I0000 00:00:1723784911.543976 160540 service.cc:154] StreamExecutor device (3): Tesla T4, Compute Capability 7.5 137/1875 ━━━━━━━━━━━━━━━━━━━━ 1s 1ms/step - loss: 0.1008 I0000 00:00:1723784912.197500 160540 device_compiler.h:188] Compiled cluster using XLA! This line is logged at most once for the lifetime of the process. 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 5s 2ms/step - loss: 0.0402 - val_loss: 0.0134 Epoch 2/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - loss: 0.0124 - val_loss: 0.0106 Epoch 3/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - loss: 0.0104 - val_loss: 0.0098 Epoch 4/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - loss: 0.0097 - val_loss: 0.0095 Epoch 5/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - loss: 0.0093 - val_loss: 0.0092 Epoch 6/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - loss: 0.0092 - val_loss: 0.0091 Epoch 7/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - loss: 0.0090 - val_loss: 0.0090 Epoch 8/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - loss: 0.0089 - val_loss: 0.0089 Epoch 9/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - loss: 0.0089 - val_loss: 0.0089 Epoch 10/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - loss: 0.0088 - val_loss: 0.0090 <keras.src.callbacks.history.History at 0x7f8ac42a3460>
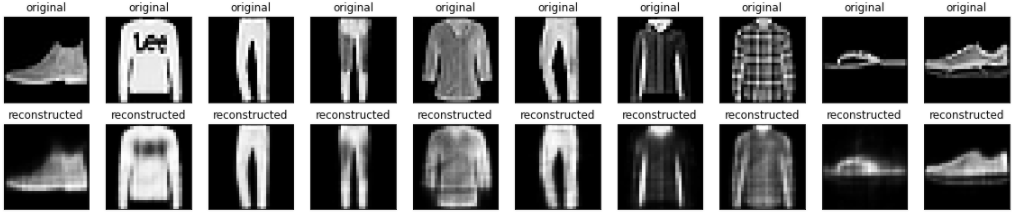
Now that the model is trained, let's test it by encoding and decoding images from the test set.
encoded_imgs = autoencoder.encoder(x_test).numpy()
decoded_imgs = autoencoder.decoder(encoded_imgs).numpy()
n = 10
plt.figure(figsize=(20, 4))
for i in range(n):
# display original
ax = plt.subplot(2, n, i + 1)
plt.imshow(x_test[i])
plt.title("original")
plt.gray()
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
# display reconstruction
ax = plt.subplot(2, n, i + 1 + n)
plt.imshow(decoded_imgs[i])
plt.title("reconstructed")
plt.gray()
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
plt.show()
Second example: Image denoising
An autoencoder can also be trained to remove noise from images. In the following section, you will create a noisy version of the Fashion MNIST dataset by applying random noise to each image. You will then train an autoencoder using the noisy image as input, and the original image as the target.
Let's reimport the dataset to omit the modifications made earlier.
(x_train, _), (x_test, _) = fashion_mnist.load_data()
x_train = x_train.astype('float32') / 255.
x_test = x_test.astype('float32') / 255.
x_train = x_train[..., tf.newaxis]
x_test = x_test[..., tf.newaxis]
print(x_train.shape)
(60000, 28, 28, 1)
Adding random noise to the images
noise_factor = 0.2
x_train_noisy = x_train + noise_factor * tf.random.normal(shape=x_train.shape)
x_test_noisy = x_test + noise_factor * tf.random.normal(shape=x_test.shape)
x_train_noisy = tf.clip_by_value(x_train_noisy, clip_value_min=0., clip_value_max=1.)
x_test_noisy = tf.clip_by_value(x_test_noisy, clip_value_min=0., clip_value_max=1.)
Plot the noisy images.
n = 10
plt.figure(figsize=(20, 2))
for i in range(n):
ax = plt.subplot(1, n, i + 1)
plt.title("original + noise")
plt.imshow(tf.squeeze(x_test_noisy[i]))
plt.gray()
plt.show()
Define a convolutional autoencoder
In this example, you will train a convolutional autoencoder using Conv2D layers in the encoder, and Conv2DTranspose layers in the decoder.
class Denoise(Model):
def __init__(self):
super(Denoise, self).__init__()
self.encoder = tf.keras.Sequential([
layers.Input(shape=(28, 28, 1)),
layers.Conv2D(16, (3, 3), activation='relu', padding='same', strides=2),
layers.Conv2D(8, (3, 3), activation='relu', padding='same', strides=2)])
self.decoder = tf.keras.Sequential([
layers.Conv2DTranspose(8, kernel_size=3, strides=2, activation='relu', padding='same'),
layers.Conv2DTranspose(16, kernel_size=3, strides=2, activation='relu', padding='same'),
layers.Conv2D(1, kernel_size=(3, 3), activation='sigmoid', padding='same')])
def call(self, x):
encoded = self.encoder(x)
decoded = self.decoder(encoded)
return decoded
autoencoder = Denoise()
autoencoder.compile(optimizer='adam', loss=losses.MeanSquaredError())
autoencoder.fit(x_train_noisy, x_train,
epochs=10,
shuffle=True,
validation_data=(x_test_noisy, x_test))
Epoch 1/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 7s 2ms/step - loss: 0.0334 - val_loss: 0.0095 Epoch 2/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 4s 2ms/step - loss: 0.0091 - val_loss: 0.0083 Epoch 3/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 4s 2ms/step - loss: 0.0082 - val_loss: 0.0078 Epoch 4/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 4s 2ms/step - loss: 0.0077 - val_loss: 0.0076 Epoch 5/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 4s 2ms/step - loss: 0.0075 - val_loss: 0.0074 Epoch 6/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 4s 2ms/step - loss: 0.0073 - val_loss: 0.0073 Epoch 7/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 4s 2ms/step - loss: 0.0072 - val_loss: 0.0072 Epoch 8/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 4s 2ms/step - loss: 0.0071 - val_loss: 0.0071 Epoch 9/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 4s 2ms/step - loss: 0.0071 - val_loss: 0.0070 Epoch 10/10 1875/1875 ━━━━━━━━━━━━━━━━━━━━ 4s 2ms/step - loss: 0.0070 - val_loss: 0.0070 <keras.src.callbacks.history.History at 0x7f8ad316bb20>
Let's take a look at a summary of the encoder. Notice how the images are downsampled from 28x28 to 7x7.
autoencoder.encoder.summary()
The decoder upsamples the images back from 7x7 to 28x28.
autoencoder.decoder.summary()
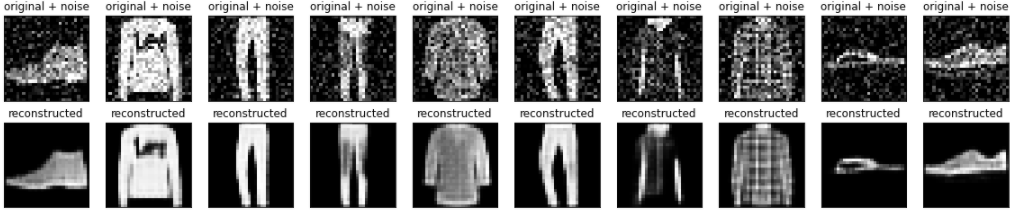
Plotting both the noisy images and the denoised images produced by the autoencoder.
encoded_imgs = autoencoder.encoder(x_test_noisy).numpy()
decoded_imgs = autoencoder.decoder(encoded_imgs).numpy()
W0000 00:00:1723784980.750910 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.770565 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.772594 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.774806 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.777004 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.780048 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.782568 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.785629 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.788932 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.791584 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.796207 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.799227 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.804537 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.809107 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.819083 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.834257 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.836833 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.839615 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.843009 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.846674 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.850582 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.856645 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.862215 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.867756 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.875814 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.883118 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.885883 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.888425 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.891329 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.894740 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.898476 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.936863 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.941096 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.944674 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.949005 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.961848 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.966838 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.973690 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.980217 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784980.999480 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.013945 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.038542 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.056058 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.077900 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.099924 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.115882 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.137481 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.186111 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.207864 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.329616 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.337779 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.352611 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.369650 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.399198 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.413427 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.443673 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.502975 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.545023 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.626188 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.642306 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.657118 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.675985 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.707544 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.718098 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.751437 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced W0000 00:00:1723784981.774578 160375 gpu_timer.cc:114] Skipping the delay kernel, measurement accuracy will be reduced
n = 10
plt.figure(figsize=(20, 4))
for i in range(n):
# display original + noise
ax = plt.subplot(2, n, i + 1)
plt.title("original + noise")
plt.imshow(tf.squeeze(x_test_noisy[i]))
plt.gray()
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
# display reconstruction
bx = plt.subplot(2, n, i + n + 1)
plt.title("reconstructed")
plt.imshow(tf.squeeze(decoded_imgs[i]))
plt.gray()
bx.get_xaxis().set_visible(False)
bx.get_yaxis().set_visible(False)
plt.show()
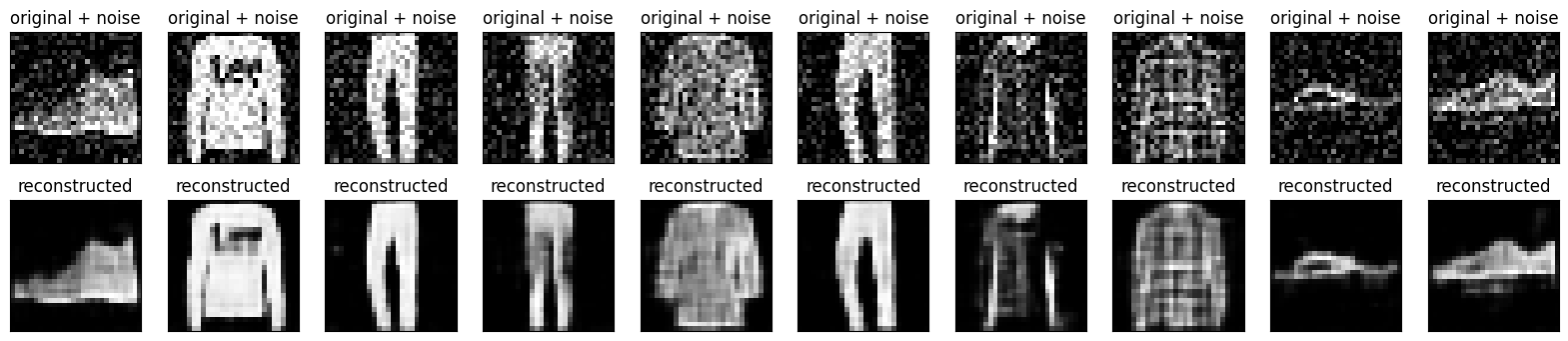
Third example: Anomaly detection
Overview
In this example, you will train an autoencoder to detect anomalies on the ECG5000 dataset. This dataset contains 5,000 Electrocardiograms, each with 140 data points. You will use a simplified version of the dataset, where each example has been labeled either 0 (corresponding to an abnormal rhythm), or 1 (corresponding to a normal rhythm). You are interested in identifying the abnormal rhythms.
How will you detect anomalies using an autoencoder? Recall that an autoencoder is trained to minimize reconstruction error. You will train an autoencoder on the normal rhythms only, then use it to reconstruct all the data. Our hypothesis is that the abnormal rhythms will have higher reconstruction error. You will then classify a rhythm as an anomaly if the reconstruction error surpasses a fixed threshold.
Load ECG data
The dataset you will use is based on one from timeseriesclassification.com.
# Download the dataset
dataframe = pd.read_csv('http://storage.googleapis.com/download.tensorflow.org/data/ecg.csv', header=None)
raw_data = dataframe.values
dataframe.head()
# The last element contains the labels
labels = raw_data[:, -1]
# The other data points are the electrocadriogram data
data = raw_data[:, 0:-1]
train_data, test_data, train_labels, test_labels = train_test_split(
data, labels, test_size=0.2, random_state=21
)
Normalize the data to [0,1].
min_val = tf.reduce_min(train_data)
max_val = tf.reduce_max(train_data)
train_data = (train_data - min_val) / (max_val - min_val)
test_data = (test_data - min_val) / (max_val - min_val)
train_data = tf.cast(train_data, tf.float32)
test_data = tf.cast(test_data, tf.float32)
You will train the autoencoder using only the normal rhythms, which are labeled in this dataset as 1. Separate the normal rhythms from the abnormal rhythms.
train_labels = train_labels.astype(bool)
test_labels = test_labels.astype(bool)
normal_train_data = train_data[train_labels]
normal_test_data = test_data[test_labels]
anomalous_train_data = train_data[~train_labels]
anomalous_test_data = test_data[~test_labels]
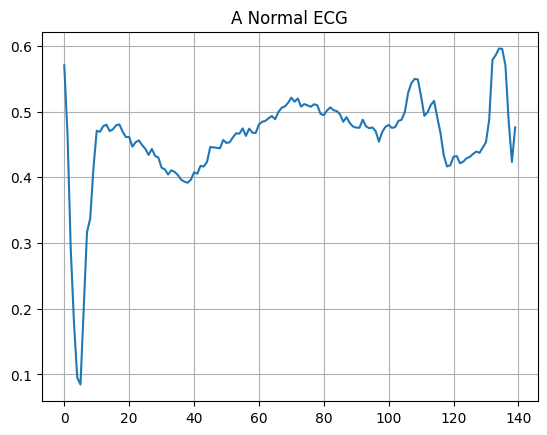
Plot a normal ECG.
plt.grid()
plt.plot(np.arange(140), normal_train_data[0])
plt.title("A Normal ECG")
plt.show()
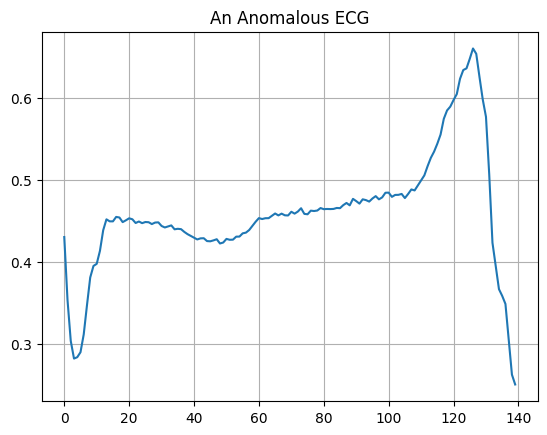
Plot an anomalous ECG.
plt.grid()
plt.plot(np.arange(140), anomalous_train_data[0])
plt.title("An Anomalous ECG")
plt.show()
Build the model
class AnomalyDetector(Model):
def __init__(self):
super(AnomalyDetector, self).__init__()
self.encoder = tf.keras.Sequential([
layers.Dense(32, activation="relu"),
layers.Dense(16, activation="relu"),
layers.Dense(8, activation="relu")])
self.decoder = tf.keras.Sequential([
layers.Dense(16, activation="relu"),
layers.Dense(32, activation="relu"),
layers.Dense(140, activation="sigmoid")])
def call(self, x):
encoded = self.encoder(x)
decoded = self.decoder(encoded)
return decoded
autoencoder = AnomalyDetector()
autoencoder.compile(optimizer='adam', loss='mae')
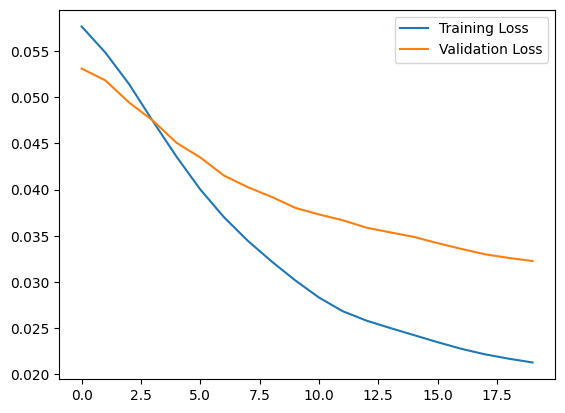
Notice that the autoencoder is trained using only the normal ECGs, but is evaluated using the full test set.
history = autoencoder.fit(normal_train_data, normal_train_data,
epochs=20,
batch_size=512,
validation_data=(test_data, test_data),
shuffle=True)
Epoch 1/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 5s 551ms/step - loss: 0.0581 - val_loss: 0.0531 Epoch 2/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0553 - val_loss: 0.0518 Epoch 3/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0519 - val_loss: 0.0494 Epoch 4/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0480 - val_loss: 0.0475 Epoch 5/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0441 - val_loss: 0.0451 Epoch 6/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0405 - val_loss: 0.0435 Epoch 7/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0374 - val_loss: 0.0415 Epoch 8/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0347 - val_loss: 0.0403 Epoch 9/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0323 - val_loss: 0.0392 Epoch 10/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0303 - val_loss: 0.0380 Epoch 11/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0286 - val_loss: 0.0373 Epoch 12/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0272 - val_loss: 0.0367 Epoch 13/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0258 - val_loss: 0.0359 Epoch 14/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0253 - val_loss: 0.0354 Epoch 15/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0244 - val_loss: 0.0349 Epoch 16/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0236 - val_loss: 0.0342 Epoch 17/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0228 - val_loss: 0.0336 Epoch 18/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0220 - val_loss: 0.0330 Epoch 19/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0215 - val_loss: 0.0326 Epoch 20/20 5/5 ━━━━━━━━━━━━━━━━━━━━ 0s 13ms/step - loss: 0.0213 - val_loss: 0.0323
plt.plot(history.history["loss"], label="Training Loss")
plt.plot(history.history["val_loss"], label="Validation Loss")
plt.legend()
<matplotlib.legend.Legend at 0x7f8ad34aa730>
You will soon classify an ECG as anomalous if the reconstruction error is greater than one standard deviation from the normal training examples. First, let's plot a normal ECG from the training set, the reconstruction after it's encoded and decoded by the autoencoder, and the reconstruction error.
encoded_data = autoencoder.encoder(normal_test_data).numpy()
decoded_data = autoencoder.decoder(encoded_data).numpy()
plt.plot(normal_test_data[0], 'b')
plt.plot(decoded_data[0], 'r')
plt.fill_between(np.arange(140), decoded_data[0], normal_test_data[0], color='lightcoral')
plt.legend(labels=["Input", "Reconstruction", "Error"])
plt.show()
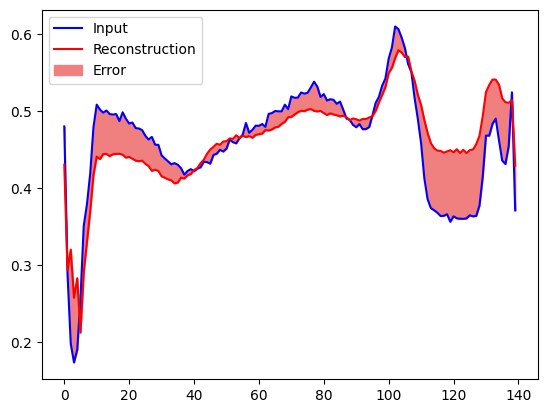
Create a similar plot, this time for an anomalous test example.
encoded_data = autoencoder.encoder(anomalous_test_data).numpy()
decoded_data = autoencoder.decoder(encoded_data).numpy()
plt.plot(anomalous_test_data[0], 'b')
plt.plot(decoded_data[0], 'r')
plt.fill_between(np.arange(140), decoded_data[0], anomalous_test_data[0], color='lightcoral')
plt.legend(labels=["Input", "Reconstruction", "Error"])
plt.show()
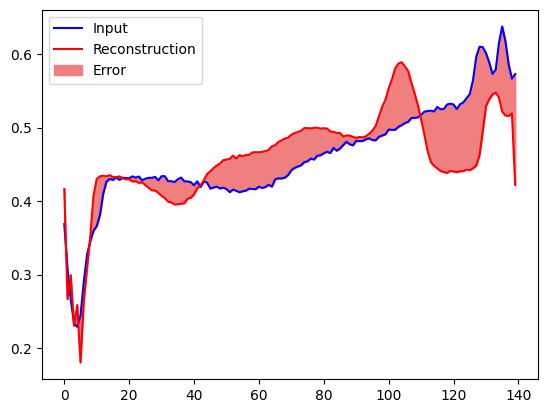
Detect anomalies
Detect anomalies by calculating whether the reconstruction loss is greater than a fixed threshold. In this tutorial, you will calculate the mean average error for normal examples from the training set, then classify future examples as anomalous if the reconstruction error is higher than one standard deviation from the training set.
Plot the reconstruction error on normal ECGs from the training set
reconstructions = autoencoder.predict(normal_train_data)
train_loss = tf.keras.losses.mae(reconstructions, normal_train_data)
plt.hist(train_loss[None,:], bins=50)
plt.xlabel("Train loss")
plt.ylabel("No of examples")
plt.show()
74/74 ━━━━━━━━━━━━━━━━━━━━ 1s 6ms/step
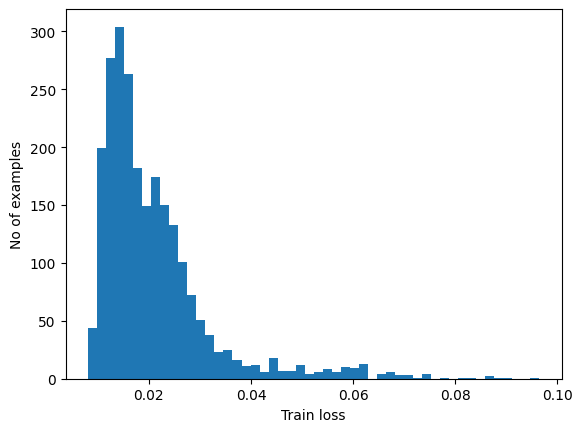
Choose a threshold value that is one standard deviations above the mean.
threshold = np.mean(train_loss) + np.std(train_loss)
print("Threshold: ", threshold)
Threshold: 0.032343477
If you examine the reconstruction error for the anomalous examples in the test set, you'll notice most have greater reconstruction error than the threshold. By varing the threshold, you can adjust the precision and recall of your classifier.
reconstructions = autoencoder.predict(anomalous_test_data)
test_loss = tf.keras.losses.mae(reconstructions, anomalous_test_data)
plt.hist(test_loss[None, :], bins=50)
plt.xlabel("Test loss")
plt.ylabel("No of examples")
plt.show()
14/14 ━━━━━━━━━━━━━━━━━━━━ 0s 19ms/step
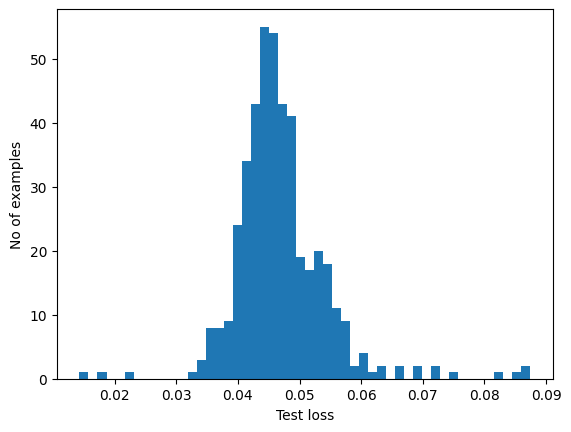
Classify an ECG as an anomaly if the reconstruction error is greater than the threshold.
def predict(model, data, threshold):
reconstructions = model(data)
loss = tf.keras.losses.mae(reconstructions, data)
return tf.math.less(loss, threshold)
def print_stats(predictions, labels):
print("Accuracy = {}".format(accuracy_score(labels, predictions)))
print("Precision = {}".format(precision_score(labels, predictions)))
print("Recall = {}".format(recall_score(labels, predictions)))
preds = predict(autoencoder, test_data, threshold)
print_stats(preds, test_labels)
Accuracy = 0.944 Precision = 0.9921875 Recall = 0.9071428571428571
Next steps
To learn more about anomaly detection with autoencoders, check out this excellent interactive example built with TensorFlow.js by Victor Dibia. For a real-world use case, you can learn how Airbus Detects Anomalies in ISS Telemetry Data using TensorFlow. To learn more about the basics, consider reading this blog post by François Chollet. For more details, check out chapter 14 from Deep Learning by Ian Goodfellow, Yoshua Bengio, and Aaron Courville.
