- Description:
The PatchCamelyon benchmark is a new and challenging image classification dataset. It consists of 327.680 color images (96 x 96px) extracted from histopathologic scans of lymph node sections. Each image is annoted with a binary label indicating presence of metastatic tissue. PCam provides a new benchmark for machine learning models: bigger than CIFAR10, smaller than Imagenet, trainable on a single GPU.
Additional Documentation: Explore on Papers With Code
Source code:
tfds.datasets.patch_camelyon.BuilderVersions:
2.0.0(default): New split API (https://tensorflow.org/datasets/splits)
Download size:
7.48 GiBDataset size:
7.06 GiBAuto-cached (documentation): No
Splits:
| Split | Examples |
|---|---|
'test' |
32,768 |
'train' |
262,144 |
'validation' |
32,768 |
- Feature structure:
FeaturesDict({
'id': Text(shape=(), dtype=string),
'image': Image(shape=(96, 96, 3), dtype=uint8),
'label': ClassLabel(shape=(), dtype=int64, num_classes=2),
})
- Feature documentation:
| Feature | Class | Shape | Dtype | Description |
|---|---|---|---|---|
| FeaturesDict | ||||
| id | Text | string | ||
| image | Image | (96, 96, 3) | uint8 | |
| label | ClassLabel | int64 |
Supervised keys (See
as_superviseddoc):('image', 'label')Figure (tfds.show_examples):
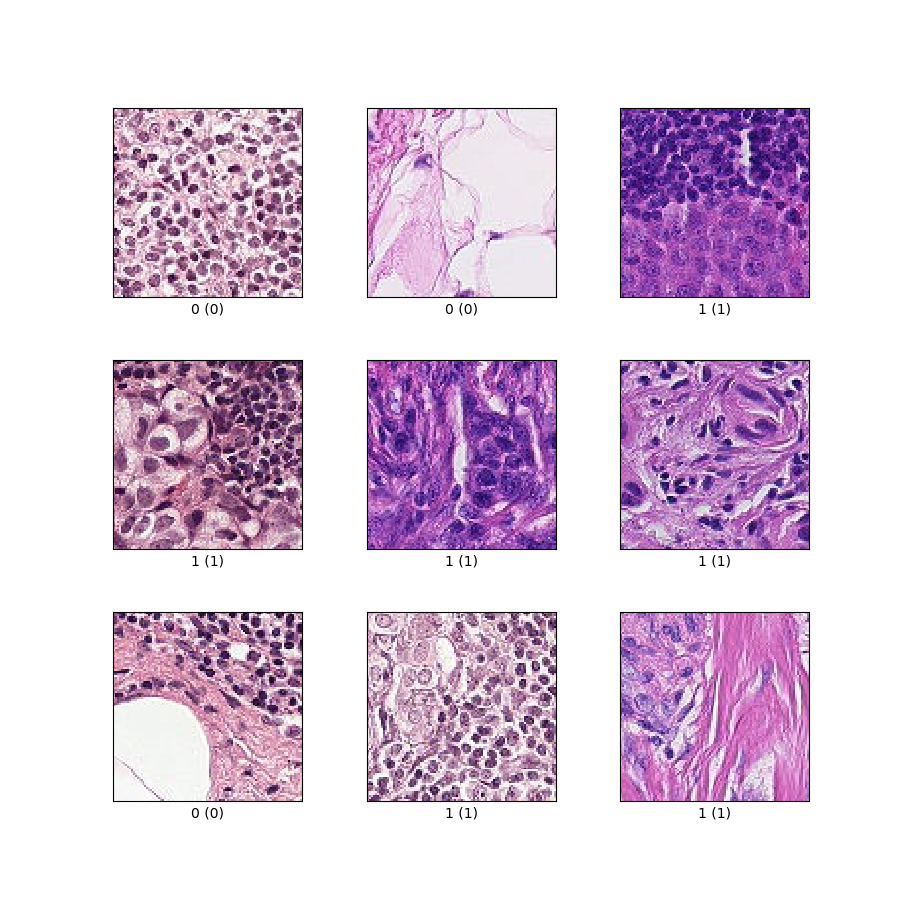
- Examples (tfds.as_dataframe):
- Citation:
@misc{b_s_veeling_j_linmans_j_winkens_t_cohen_2018_2546921,
author = {B. S. Veeling, J. Linmans, J. Winkens, T. Cohen, M. Welling},
title = {Rotation Equivariant CNNs for Digital Pathology},
month = sep,
year = 2018,
doi = {10.1007/978-3-030-00934-2_24},
url = {https://doi.org/10.1007/978-3-030-00934-2_24}
}
